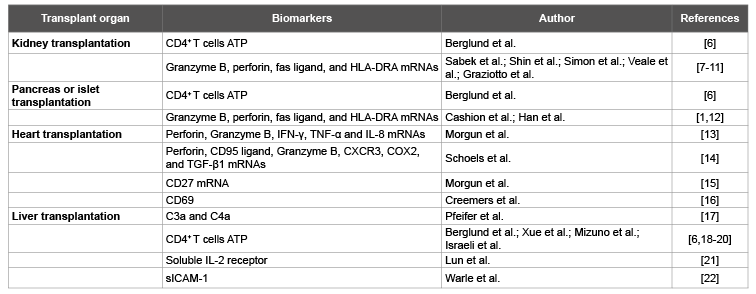
Table 1:Biomarkers of allograft rejection from peripheral blood cells.
GuoYing Wang*
Transplantation Research Institute, Sun Yat-sen University, China*Corresponding author: GuoYing Wang, Liver Transplantation Center, Third Affiliated Hospital, Transplantation Research Institute, Sun Yat-sen University, NO. 600 TianHe Road, 510630 Guangzhou, China, E-mail: wanggy3@126.com
Timely diagnosis and treatment of acute and chronic rejection after organ transplantation are very important for the improvement in allograft and patient survival. In addition, over-immune suppression increases the risk of infection, cancer, and drug side effects. Although allograft biopsy with conventional histologic examination remains the gold standard for diagnosing rejection, a non-invasive procedure that can be used as the early diagnostic tools is necessary for detection of allograft rejection considering the safety of repetitive biopsies. Emerging data suggest that development of non-invasive biomarkers applied for prediction of allograft rejection is feasible. MicroRNA-mediated RNA interference appears to play an important role in modulating the immune response. Many studies indicate that microRNAs that are selectively and/or highly expressed in immune cells have a permissive effect on the maturation, proliferation and differentiation of myeloid and lymphoid cells. In this mini review study, we summarized the immune-related microRNAs and hypothesized the predictive value of microRNAs in evaluating immune status and predicting the development of immune rejection in transplant recipients.
Organ transplantation has become the optimising approach to the treatment of failure of the heart, liver, kidney, and lung. However, the use of immunosuppressive drugs increases the risk of infection, malignancy and drug side-effects, which is associated with significant morbidity and mortality in transplant patients. Conversely, insufficient immunosuppressive drug exposure or interruption of drug therapy often increases the risk of rejection. Acute and chronic rejection events after organ transplantation, which arise at least partly because immunosuppressive regimens fail to inhibit alloimmune responses completely, have a strong impact on long-term allograft survival. It is desirable to balance the level of immunosuppression to avoid episodes of rejection, infectious diseases, malignancies and drug side-effects. Most transplant centers rely on monitoring trough levels of immunosuppressive drugs combined with specific biomarkers for allograft dysfunction (e.g., serum creatinine levels in kidney transplantation and aminotransferases levels in liver transplantation) to assess the allograft status. Although the functional parameters such as creatinine and aminotransferases suggests acute rejection occurrence, biopsy of the transplanted allograft remains the gold standard for the assessment of overall graft status and diagnosis of acute rejection. However, due to its invasive nature, this approach is of limited use frequently in many centers. Moreover, allograft biopsy diagnoses rejection at a relatively advanced stage of immune process and tissue injury, and fails to recognize subclinical rejection that is histologic abnormalities before the change of biochemical markers of graft dysfunction. Furthermore, the diagnosis of subclinical rejection or the assessment of therapeutic efficacy of anti-rejection requires repeated biopsies. The biopsy can result in complications, patient discomfort and sampling errors may bias the histological diagnosis. Not uncommonly, some patients received empirical anti-rejection therapy for presumed acute rejection prior to the availability of a confirmatory histological report, which may present a diagnostic dilemma for the transplant clinicians. Thus, there is an urgent need for specific and sensitive noninvasive biomarkers for early diagnosis of acute rejection.
Development of predictive and diagnostic biomarkers of allograft status is important, and may be beneficial for efficient individualized therapy in the recipients [1-5]. An individualized immunosuppressive protocol based on immune monitoring would be useful for avoiding undesired effects. However, until now, there has been no simple and effective method available for the direct assessment of immune status in transplant recipients. Changes of gene or protein expression pattern of non-invasively collect biological samples (e.g., blood, urine, etc.) have been investigated as biomarkers of allograft status. Non-invasive procedures have several advantages including sequential and repeated assessments of immune status of transplant recipients. Molecular perturbations or altered protein expression may precede not only graft dysfunction but also histological changes. More importantly, these non-invasive measurements may serve to guide minimization of maintenance immunosuppression and individualization of immunosuppressive therapy. Transplant clinicians can more accurately monitor the immune status of transplant patients and may be able to promptly adjust the immunosuppressive therapy. A noninvasive approach based on molecular markers could not only serve as a surrogate tool instead of the invasive biopsy procedure, but could also provide predictive and diagnostic information about allografts with early subclinical rejection.
The rapid developments of immunology and molecular biology have led to a deep understanding of the anti-allograft immune response and have informed biomarker development. Numerous blood-based noninvasive tests for acute rejection have been investigated using T cell activation markers as the parameters (Table 1) [6-22]. Gene products associated with the immune response have been evaluated as biomarkers of allograft status. For example, gene expression signatures of peripheral blood cells have been evaluated as non-invasive biomarkers of kidney transplant dysfunction [5].

Table 1:Biomarkers of allograft rejection from peripheral blood cells.
Management of organ transplant rejection by an immune cell function assay to assess the immune status of the transplant patients and to individualize immunosuppressive therapy has been proposed. The ImmuKnow assay was developed to serve as a useful tool to assess allograft status and adjust the immunosuppressive treatment [6,18-20]. It has also been investigated as a method of predicting early allograft rejection after liver or kidney transplantation [20,23,24]. However, there is insufficient evidence of the effectiveness of the ImmuKnow assay in the management of organ transplant rejection in individuals undergoing immunosuppressive therapy after organ transplant and for the identification of individual risk for developing rejection prior to allograft biopsies. The transplant patients being highly over immunosuppressed as assessed by the ImmuKnow assay do not seem to have a lower risk of short-term rejection [6]. Nucleic acidbased biomarkers of allograft status and outcome such as messenger RNA profiles have already been reported by several transplant centers [2,3,25].
MicroRNAs (miRNA) are small noncoding RNAs, which are encoded in a highly conserved part of genomic clusters. Fully functional mature miRNA requires cytoplasmic processing by an RNase III enzyme, Dicer, producing a [19-25] nucleotide product, capable of being incorporated into the RNA-induced silencing complex (RISC). The RISC can recognize complementary mRNA transcripts for degradation or translational silencing. Each miRNA can regulate one to several mRNA transcripts, and conversely one mRNA can be regulated by one to several miRNA. It is estimated that at least half of mRNA may be regulated by miRNA [26]. Many studies have demonstrated that miRNA as a new immuneregulatory agent plays an important role in the regulation of innate and adaptive immune responses [27]. It is found that a wide range of miRNA are involved in the regulation of immunity, including the development and differentiation of B and T cells, proliferation of monocytes and neutrophils, antibody switching and the release of inflammatory mediators, and their cells at different stages of differentiation have distinct miRNA expression profiles [27-29]. MiRNA-146a as a negative regulator of the innate immune response and a central role for miR-155 in the regulation of T and B cell responses during the acquired immune response have emerged from many studies [27]. MiRNA modulate the induction, function and maintenance of the regulatory T cells [30]. MiRNA are also important for regulating the differentiation of dendritic cells and macrophages [31]. Through the modulation of transcription and translation, the expression of specific sets of miRNA has emerged as a critical regulatory principle in mammalian immune system. Deregulation of certain individual miRNA, which modulates protein expression of downstream hundreds of target genes, may lead to reduced tolerance against self-antigens and the development of immune disorders like autoimmunity and cancer. Moreover, deregulation of miRNA fine-tuning the immune response may lead to sustained activation of inflammation and immune responses.
Today, organ transplantation is the only treatment available for patients with end-stage organ failure. However, immunosuppressive therapy remains non-specific and has tremendous negative consequences upon allograft and patient survival. Clinical immunosuppressants lead often to over-or under-immunosuppressed organ recipients, which in turn increase the risk of major complications such as infection or chronic rejection. In order to accurately balance the immune response, a wide range of potential specific markers are assessed in regard of their prediction potential related to acute rejection and graft outcome [2,32]. Recent studies have demonstrated that the expression of specific miRNA as a critical regulatory mechanism play an important role in the immune system. Here, we hypothesize that miRNA expression profiles in peripheral blood may predict the development of acute and chronic rejection, minimize the need for invasive biopsy procedures, and facilitate personalization of immunosuppressive therapy for the allograft recipient. The analysis of differential miRNA expression profiles in peripheral blood may prove useful in reducing the potential risks of rejection, infection and other complications following transplant.
A number of studies have demonstrated the role of miRNA in physiologic and pathologic circumstances such as inflammatory, infectious diseases and cancer. Anglicheau et al. [33] compared the levels of miRNA expression in renal biopsies of acute rejection after renal transplantation between the normal samples, and try to reveal the relation between miRNA and acute rejection after renal transplantation. The results have showed that the expression of miRNA as a new immuneregulatory agent in peripheral blood and allograft has a strong association with acute rejection after renal transplantation. The ImmuKnow assay, a measure of intracellular CD4+ T cell ATP release in transplant recipients, is a non-invasive means of rapidly providing a biomarker of patient cellular immune status, which is considered as a potential tool to identify patients at risk for opportunistic infections or acute rejection [6,18-20]. However, The ImmuKnow assay needs to use special testing tools. Together with existing clinical tools, miRNA expression assay may provide a better assessment of immune status in transplant recipients. Further studies are in progress to identify the specific miRNA which close relationship with immune status of transplant patients. A multiparametric immune monitoring program, performed frequently and lifelong, might be the appropriate approach to predict dysfunction of the graft before clinical symptoms occur.
Measurements of microRNA expression may offer a particularly useful diagnostic tool to monitor allograft viability or to detect organ rejection. Non-invasive approach such as mRNA and miRNA for predicting the development of an episode of acute rejection would be as a substitute for the invasive allograft biopsy procedure and for prognosticating allograft outcome. We hypothesize that miRNA expression patterns would facilitate in the near future individualization of immunosuppressive therapy in organ graft recipients. The developments in the area of mechanism based non-invasive tests for the diagnosis of acute rejection of solid organ allografts will facilitate a better assessment of immune status in transplant recipients. With technological developments of functional genomics and proteomics of allograft rejection, it is reasonable to further develop specific and sensitive non-invasive tests for acute rejection that provide mechanistic insights as well as help individualize therapy.
This work was supported by grants from the National Natural Science Foundation of China (No. 81370575, 81470870, 81370555, 81372243), Science and Technology Program of Guangzhou (2014J4100183, 201400000001-3), Sci-tech Research Development Program of Guangdong Province (2014A020211015), Natural Science Foundation of Guangdong Province (2015A030312013, 2015A030313038).
Download Provisional PDF Here
Article Type: Opinion Article
Citation: Wang GY (2016) Differential microRNA Expression in Peripheral Blood as a useful Predictor for Immune Status after Solid Organ Transplantation. J Gastric Disord Ther 2(2): doi http://dx.doi. org/10.16966/2381-8689.116
Copyright: © 2016 Wang GY. This is an openaccess article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Publication history:
All Sci Forschen Journals are Open Access