Introduction
Applications of therapeutic genes and siRNAs have tremendously increased due to their huge potential in clinical medicine. Gene therapy involves delivery of a therapeutic gene, enabling it to transcribe and translate into protein of interest, or a nucleic acid sequence to silence the expression of a specific gene, within a particular cell type. Genetic manipulation of human cells through transportation of functional genes such as plasmid DNA (pDNA) and siRNA is an attractive approach to treat many critical genetic and acquired diseases having single or multiple gene defects [1]. However, naked therapeutic genes and siRNAs are rapidly degraded and inefficient for cellular uptake with poor transfection activity and gene-knockdown effectivity, respectively. Development of ideal gene carriers is therefore fundamental to the success of gene therapy with acceptable safety profiles [2].
Intensive efforts in the last four decades have led to the development of a number of vehicles for nucleic acid delivery, generally classified into viral and non-viral vectors [3]. Although the viral vectors are highly effective in overcoming the multiple extracellular and intracellular barriers commonly observed in the delivery process, they possess critical limitations including immunogenicity, carcinogenicity and low DNA carrying capacity, restricting their applications in clinical medicine [4]. On the contrary, non-viral vectors, such as cationic lipids, polymers, lipids and peptides, and inorganic nanoparticles are relatively safe, but possess limitations in terms of poor physical stability, quick blood clearance by reticuloendothelial system, low cellular internalization, inefficient endosomal escape and poor nuclear translocation [5,6]. Improving the transfection efficacy of non-viral vectors is of crucial importance for successful gene therapy in the future.
Inorganic nanoparticles constitute an important branch of non-viral vectors. Unlike their organic counterparts, inorganic vectors demonstrate several distinctive features, such as well- defined particle dimension, stability and, unique magnetic and electrical properties. Inorganic nanoparticles directed to transport the nucleic acids to the targeted cells focusing on pre-clinical studies were gold, silica, carbonate apatite, carbon nanotubes, quantum dots and iron oxides [7-13]. However, most of the inorganic nanoparticles currently being investigated are not biodegradable, raising great concerns in their clinical implications. In addition, surface-modification is commonly required to prevent their selfaggregation and enable them to interact with nucleic acids, surface-coating agents or cell-recognizable ligands [14-17]. Precipitation reaction is one of the new approaches in synthesizing nanoscale inorganic materials, exploiting many unique and interesting physical and chemical properties of insoluble salts. It involves a reaction between two soluble salt reactants in an aqueous solution to form an insoluble salt product via induction of super saturation [18,19]. Generation of such salt nano-precipitates requires the involvement of simple equipment and offers the flexibility in controlling particle sizes and compositions in near ambient temperature and pressure. Recent development has shown favorable outcomes from the nanoparticles prepared from precipitation method, including calcium carbonate (CaCO3), iron oxide (Fe3O4) and barium sulfate (BaSO4 ) [20-22].
Barium compounds have been associated with many applications in medical field. Insoluble barium compounds are mostly non-toxic, allowing them to be used in clinical and cosmeceutical areas [23]. BaSO4 , for instance, with low toxicity and high opacity for X-ray imaging is used as a radio contrast agent for digestive tract. High solubility of barium salts in acidic environment might also help release pDNA and siRNA from the particles into endosome, therefore promoting the subsequent steps of their processing for transgene expression and silencing of a target endogenous gene, respectively [22]. In this study, nanocrystals formed from various barium formulations have been tested for their potential ability to adsorb pDNA and siRNA, transport these nucleic acids into 4T1 and MCF-7 cells, facilitate silencing of target protein expression after intracellular delivery MAPK siRNA and induce cytotoxicity following delivery of MAPK siRNA and p53 gene into the breast cancer cells.
Materials and Methods
Reagents
Barium chloride (BaCl2 ), Sodium sulfate (Na2SO4 ), Sodium sulfite (Na2SO3 ), Sodium fluoride (NaF), Sodium carbonate (Na2CO3 ), Sodium phosphate (Na2HPO4 ), 4-(2-hydroxyethyl)- 1-piperazineethanesulfonic acid (HEPES), Ethidium bromide (EtBr) and Propidium iodide (PI) were obtained from Sigma-Aldrich (Missouri, USA). Dulbecco’s Modified Eagle Medium (DMEM) was purchased from Gibco BRL (Tennessee, USA). Plasmid pGL3 (Promega, USA) containing luciferase gene under SV40 promoter and plasmid pEGFP-N1 containing green fluorescence protein (pGFP) gene were extracted from DH5α bacteria strain of Escherichia coli (E.coli) with Qiagen plasmid extraction kit (Hilden, Germany). AF 488 negative siRNA (fluorescence siRNA) was acquired from Qiagen, Germany.
Cell culture
MCF-7 and 4T1 cells were cultured in 75 cm2 flasks in DMEM supplemented with 10% fetal bovine serum (FBS) (Sigma Aldrich, USA), 10% HEPES and 50 µg of penicillin ml-1 at 37°C in a humidified 5% CO2 - containing atmosphere. When the cells were close to 90% confluency, subculturing was performed following trypsinization of the adherent cells and pouring the detached cells in appropriate number in a new plate with 10% serum-supplemented DMEM.
Fabrication and screening of potential barium salt crystals
Each type of barium salt particles was prepared by incorporating 5 µl of 1 M of cation- providing BaCl2 salt into 10 µl HEPES-buffered solution (pH adjusted to 7.5) and mixing the solution with 5 µl of 1 M of one anion-providing salt, Na2SO4 , Na2SO3 , NaF, Na2CO3 or Na2HPO4 . The final mixture was incubated for 30 min at 37°C and subsequently added to 10% FBS-supplemented DMEM medium to obtain the final volume of 1 ml particle suspension. Absorbance at 320 nm wavelength was measured for all fabricated barium salt and carbonate apatite (CO3 AP) particles spectrophotometrically (UV 1800 Spectrophotometer, Shimadzu, Japan). Preparation of CO3 AP nanoparticles involves dissolving exogenous CaCl2 at 5 mM with 1 ml of bicarbonate-buffered DMEM (pH 7.4) and incubation for 30 min at 37°C, followed by addition of 10% FBS to the generated CO3 AP particles to prevent further growth or aggregation of the particles. The spectrophotometric measurement was done in triplicates and the data were plotted into a graph with mean ± SD.
Manipulation of concentrations of reacting salts for generation of nanoscale particles was performed through addition of BaCl2 ranging from 2 µl to 10 µl of 1M into HEPES buffer solution with subsequent mixing of 5 µl of 1M Na2SO4, Na2SO3 , NaF, Na2CO3 or Na2HPO4 with the resultant solution and incubation for 30 minutes at 37°C. Time-dependent analysis of particle formation was made by addition of 5 µl of 1M BaCl2 to 10 µl HEPES-buffered media (pH 7.5) and mixing with 2 µl of 1 M Na2SO4 , Na2SO3 , NaF, Na2CO3 or Na2HPO4 , followed incubation at 37°C for 0, 30, and 60 minutes and subsequent mixing of serum-supplemented DMEM. The effect of pH on particle formulation was analyzed through preparation of HEPES-buffered solutions of various pHs ranging from 4 to 9, and mixing of 5 µl of 1 M BaCl2 with 2 µl of 1 M Na2SO4 , Na2SO3 , NaF, Na2CO3 or Na2HPO4 in the solutions prior to incubation at 37°C for 30 min. The influence of changes in incubation temperature was explored through incubation for 30 min at various temperatures (4°C, 37°C and 70°C) of the generated salt crystals following mixing of 5 µl of 1M BaCl2 and 2 µl of 1 M Na2SO4 , Na2SO3 , NaF, Na2CO3 or Na2HPO4 in the HEPES buffer of a fixed pH (7.4) and incubation time. CO3 AP was included in each experiment as a positive control. Experimental studies were completed in triplicates and analysis was presented as graphs with mean ± SD.
Size estimation and zeta potential measurement of NPs
Size and zeta potential measurement of fabricated nanoparticles utilizing Zeta Sizer (Malvern, Nano ZS, UK) was performed after mixing of 5 µl of 1 M BaCl2 and 2 µl to 10 µl of 1 M Na2SO4 , Na2SO3 , NaF, Na2CO3 or Na2PO4 in a HEPES buffer, followed by incubation at 37°C for 30 minutes. The generated salt crystals were maintained on ice prior to measurement with Zetasizer. A refractive index (RI) ratio of 1.325 (measured with DMEM media by refractometer) was used to calculate particle sizes and zeta potential. Analysis of data was carried out using Zetasizer software 6.20 and all salt samples were measured in duplicate and shown as mean ± SD. The size and morphology of various nanocrystal samples were visualized through Field Emission scanning electron microscopic (Hitachi S-4700 FE-SEM, Japan). Nanoparticles were centrifuged at 15,000 RPM for 10 seconds, followed by removal of supernatant and resuspension with milli- Q water. Salt particle suspensions were maintained under ice before microscopic observation. 1 µl of each sample was placed onto carbon tapecoated sample holder and dried at room temperature, followed by platinum sputtering of the dried samples for 30 seconds. Microscopic observation of the sputtered nanoparticles was done using FE-SEM at 10-15 kV.
Binding affinity of pDNA and siRNA towards particles
A study involving binding affinity of pDNA and siRNA towards various barium salt particles includes qualitative and quantitative measurement of the fluorescence-labeled pDNA and siRNA associated with the particles. 1 µg pDNA (pGFP) was labeled non-covalently with propidium iodide (PI) at 1:1 ratio and added to 5 µl of 1 M BaCl2 in 10 µl HEPES-buffered solution, followed by incorporation of 2 µl of 1 M Na2SO4 , Na2SO3 , NaF, Na2CO3 or Na2HPO4 into the solution to generate respective salt precipitates carrying fluorescent pDNA, through incubation at 37°C for 30 minutes. DMEM medium was added to form the final volume of 1 ml particle suspension. CO3 AP particles (positive control) were made at pGFP: PI ratio of 1 according to same procedure as described above. Microscopic visualization of the aggregated particles was achieved following addition of the fabricated salt crystals to each well of a 24-well plate (Nunc, Denmark), and observing under a fluorescence microscope (Olympus, Japan). Quantitative measurement of nanoparticles-bound pDNA involved multi-label plate reading of the supernatant representing the unbound fraction of DNA (Victor X5, Perkin Elmer), following centrifugation of the differentially formulated particles at 15,000 RPM for 5 minutes. The 100 µl supernatant was aspirated and transferred into 96- well plate (Nunc, Denmark), prior to fluorescence intensity measurement.
Binding affinity of siRNA for particles was determined following inclusion of 100 nM AF 488 negative control siRNA (fluorescence siRNA, Qiagen, Germany) into 5 µl of 1M BaCl2 and subsequent addition of 10 µl HEPES-buffered solution along with 2 µl of 1 M Na2SO4 , Na2SO3 , NaF, Na2CO3 or Na2HPO4 , prior to incubation at 37°C for 30 minutes. Each experiment was done in triplicates and analyzed with mean ± SD.
Cellular uptake of particles with bound pDNA and siRNA
MCF-7 cells from exponentially growth phase were seeded at 50,000 cells per well into 24-well plates the day before transfection. Cellular uptake of particle-bound siRNA was studied by introduction of 10 nM of AF 488 siRNA (Qiagen, Germany) to 5 µl of 1M BaCl2 , followed by mixing with 10 µl HEPES buffer and 2 µl of 1 M Na2SO4 , Na2SO3 , NaF, Na2CO3 or Na2HPO4 , and incubation at 37°C for 30 minutes to form the particle-siRNA complexes. DMEM media was added to the precipitates to form 1 ml final volume of particle suspension. 10% FBS was finally added to the suspension. Salt particles of each type were incubated with seeded carcinoma cells for 4 hours, prior to removal of particle suspension, washing of the cells with 10 mM EDTA in PBS and addition of 100 µl of serum-supplemented medium.
Determination of cytotoxic effects of particles
Cytotoxicity of barium salt particles was determined by cell viability (MTT) assay, following incubation of treated cells for 24 to 72 hours. 5 µl of 1 M BaCl2 was mixed with 2 µl of 1 M Na2SO4 , Na2SO3 , NaF, Na2CO3 or Na2 HPO4 without nucleic acid in 10 µl HEPES-buffered solution. After 30 min incubation at 37°C, the generated particles were exposed to MCF-7 and 4T1 cells for 1-3 days, with CO3 AP nanoparticles included as a control. The fraction of the viable cells was determined using MTT assay. Briefly, 50 µL of MTT (5 mg/ml in PBS) was added aseptically to the treated cells in each of the wells, followed by incubation at 37°C and 5% CO2 for 4 hours. Medium containing MTT was aspirated after the incubation period and the formazan crystals formed at the bottom of each well were dissolved by addition of 300 µL dimethyl sulfoxide (DMSO) solution. Absorbance of the resulting formazan solution was determined spectrophotometrically at 595 nm wavelength using microplate reader (Dynex Opsys MR, USA) with reference to 630 nm.
Evaluation of gene expression and silencing efficacy in vitro
1 ml suspension of each type of salt particles loaded with pGFP or luciferase reporter vector (pGL3) or p53 gene-carrying plasmid and supplemented with DMEM media, was introduced into each well with approximately 50,000 MCF-7 or 4T1 cells seeded in the previous day. 1 µg pGFP, pGL3 or p53 was mixed with 5 µl of 1 M BaCl2 before addition of 2 µl of 1 M Na2SO4 , Na2SO3 , NaF, Na2CO3 or Na2HPO4 to generate respective salt precipitates in 10 µl HEPES solution through incubation at 37°C for 30 min. Serum-supplemented DMEM medium was added to achieve 1ml of salt suspensions. CO3 AP was prepared following addition of pDNA and 5 mM exogenous CaCl2 into prepared 1 ml DMEM media, and incubation at 37°C for 30 min, prior to addition of 10% FBS to the suspension. The cells were treated with various formulations for a consecutive period of 4 hrs, before removal of the particle-containing media and brief washing of the treated cells with 10 mM EDTA in 1X PBS. Upon substitution with 1 ml serum-containing DMEM media, the cells were incubated for 48 hrs, prior to observation of gene expression through fluorescence microscopy (pGFP) and luciferase reporter assay (pGL3) using a commercial kit (Promega, USA) and photon counting (Beckman Coulter, USA). Quantitative luciferase assay was repeated thrice and expressed in a graph as mean ± SD of luminescence activity/mg of protein.
The effect of p53 gene expression on cell viability was evaluated by addition of 50 µl of MTT (5 mg/ml in 1X PBS) solution to the cells treated at first for 4 hrs with p53 plasmid-bound particles, washed with 5 mM EDTA in PBS and further incubated in fresh serum medium for 48 hrs. Medium containing MTT was aspirated post 4 hrs incubation, with formed formazan crystals in each well dissolved by mixing with 300 µl DMSO solution. Medium containing naked siRNA represented the negative control for the study. Quantitative measurement of formazan crystals in the form of optical density (OD) was done at 595 nm wavelength with reference to 630 nm using microplate spectrophotometer (Bio-Rad, USA).
To examine the effect of knockdown of an endogenous gene and expression/activation levels of the associated signaling molecules following intracellular delivery of a particular siRNA using various barium salt particles, 10 nM MAPK siRNA was introduced into 5 µl of 1M BaCl2 , followed by incorporation of 2 µl of 1 M Na2SO4 , Na2SO3 , NaF, Na2CO3 or Na2HPO4 in 10 µl HEPES media, and incubation at 37°C for 30 min. Serum-supplemented DMEM media was used to top up the particle suspension to 1 ml. MAPK siRNA-loaded CO3 AP (positive control) was prepared by addition of MAPK siRNA and 5mM exogenous CaCl2 to 1 ml of DMEM medium and incubation at 37°C for 30 min. Finally, 10% FBS as added to the suspension. Cells were treated at first for 4 hrs, followed by removal of medium, washing with EDTA in PBS and substitution with 1 ml serum- supplemented DMEM medium. Subsequent incubation was performed for 48 hrs, before addition of 50 µl of MTT to the cells in each well to form formazan crystals by metabolically active cells. Medium containing MTT was aspirated post 4 hrs incubation, with the formed formazan crystals in each well dissolved by mixing with 300 µl DMSO solution. Medium containing only siRNA (no salt) represented the negative control for the study. Quantitative measurement of formazan crystals in the form of OD was done at 595 nm wavelength with reference to 630 nm using microplate spectrophotometer (Biorad, USA). The cell viability of siRNA-loaded particles and naked siRNA (without NPs) was calculated based on the equation:
\[\% {\rm{ }}cell{\rm{ }}viability:{\rm{ }}\frac{{OD{\rm{ }}loaded{\rm{ }}NCs{\rm{ }}--{\rm{ }}OD{\rm{ }}reference}}{{OD{\rm{ }}naked{\rm{ }}siRNA{\rm{ }}--{\rm{ }}OD{\rm{ }}reference}} \times {\rm{ }}100\]
Each experiment was carried out in triplicates and expressed in graph as mean ± SD of cell viability.
Cells treated with MAPK-siRNA in free and particle-bound forms (BaSO4 , BaSO3 and BaF2) were individually lysed with IP lysis buffer and subjected to centrifugation at 13,000 RPM for 20 min at 4°C. Supernatant comprising protein sample was collected and 5 µl was aspirated to estimate the total amount of proteins through bovine serum albumin (BSA) assay kit based on the manual. In the initial step, BSA protein was used to create the standard curve, which was used to calculate the total protein concentration of cellular lysates based on their absorbance intensity. The remaining samples were aliquoted and stored in -80°C for subsequent SDS-PAGE and Western blot. The cellular lysates containing 30 µg of total protein were mixed with 10 µl of 10X loading dye and subjected to SDSPAGE using stain free mini protein SFX gels (15 wells) in 1X running buffer at 0.01 amp/gel. 7 µl precision plus protein standards-dual color was used as molecular weight marker to establish the molecular weight of the sampled proteins. Transfer of protein samples from gel to the 0.2 µm PVDF membranes attached to trans-blot turbo transfer pack through trans-blot turbo transfer system was performed for 7 minutes at 1.3 amp, followed by blocking in 5% skimmed milk in 1X TBST for 1 hour at room temperature. The membrane was next incubated with primary antibodies (pMAPK, TMAPK, pAKT, TAKT and GAPDH as loading reference) at 4°C overnight with gentle shaking, followed by washing in 1X TBST for 5 times to remove unbound primary antibodies. HRP-conjugated goat anti-rabbit secondary antibody Ig G (1:3000) was introduced into the membrane for 1 hour with mild agitation, before washing for 5 times in 1X TBST to again eliminate the unbound antibodies. The membrane was exposed to mixture of ECL for 5 minutes before observation of bands through chemiluminescence signals using XRS Chemidoc system (BioRad, USA).
Results and Discussions
Generation of salt precipitates
Direct mixing of BaCl2 with Na2SO4 , Na2SO3 , NaF, Na2CO3 or Na2 HPO4 , followed by incubation at 37°C for 30 min resulted in microscopically visible particles. The formation of particles is normally accompanied by growth and aggregation of the particles, providing a sharp increase in absorbance at 320 nm as a reflection of turbidity (Figure 1A) and demonstrating large- sized crystals under optical microscope (Figure 1B). Optimization of pH, concentrations of reactants and time and temperature of incubation is therefore vital in fabricating effective nanoparticles. Particle formation accelerates as the concentration of reactants increases, thus acting as a driving force for the chemical reaction [24] (Figure 1).
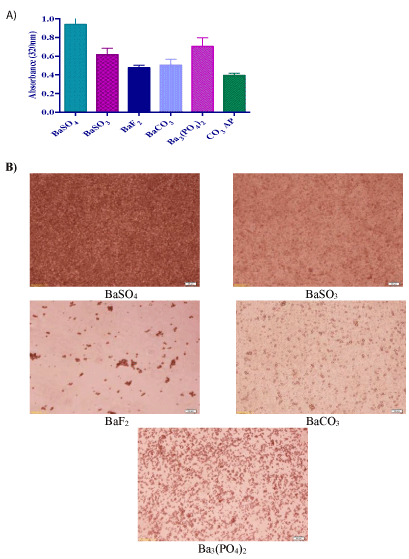
Figure 1: Turbidity measurement and optical microscopic images of barium salt particles. (A) Absorbance intensity of the particles formed by precipitation reaction. Addition of 5 µl of 1 M BaCl2 , into 10 µl HEPES buffered media (pH adjusted to 7.5), followed by mixing of 5 µl of 1 M Na2SO4 , Na2SO3 , NaF, Na2CO3 or Na2HPO4 , generated various salt crystals upon 30 min incubation at 37°C. Subsequently, serum-supplemented DMEM media was added to achieve 1 ml particle suspension. Absorbance at 320 nm wavelength was measured for all fabricated particles using a spectrophotometer with reference to CO3 AP. (B) Microscopic observation of particles formed by precipitation reaction. Images were captured at 10X resolution, with reference to (f) CO3 AP.
Investigation on the effect of reactant concentrations based on turbidity measurement (Figure 2A) and optical microscopic images (Figure 2B) showed that particle formulation with 2 mM of barium salts exhibited reduced particle growth and less aggregates compared with more concentrated salts, suggesting that particle growth is dependent on reactant concentrations when time and temperature of incubation and pH are fixed. As shown in figure 2A, with an increase in the cation-providing salt, BaCl2 (from 2 µl, to 10 µl of 1 M) with a constant amount (5 µl of 1 M) of an anion-providing salt, Na2SO4 , Na2SO3 , NaF, Na2CO3 or Na2HPO4 , the rate of precipitation reaction was enhanced with the result of increased absorbance or turbidity. Similarly, the rate of particle synthesis was accelerated with increasing concentrations (2 µl, to 10 µl of 1 M) of an anion-providing salt (Na2SO4 , Na2SO3 , NaF, Na2CO3 or Na2HPO4 ) in presence of a fixed concentration of BaCl2 (cation-providing salt) (Figure 2B). The nucleic acid binding efficiency of a carrier is regulated by particle size and number, and available charges, which in excess might may lead to generation of huge aggregates, preventing optimum formation of DNAparticle complexes and hindering cellular uptake of the complexes [25] (Figure 2).
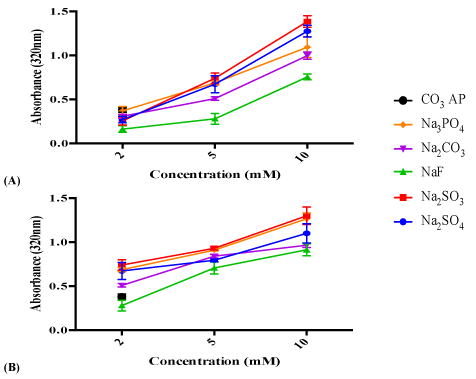
Figure 2: (A) Effect of cation-providing salt concentration on particle formation. Various concentrations of (a) BaCl2 was introduced (2 µl, 5 µl and 10 µl of 1M) into 10 µl HEPES buffered media (pH 7.5), followed by mixing of 5 µl of 1 M Na2SO4 , Na2SO3 , NaF, Na2CO3 or Na2HPO4 , generating various salt particles upon 30 min incubation at 37°C. (B) Effect of anion-providing salt concentration on particle formation. 5 µl of 1 M BaCl2 was introduced into 10 µl HEPES buffered media (pH adjusted to 7.5), followed by mixing of different concentration of Na2SO4 , Na2SO3 , NaF, Na2CO3 or Na2HPO4 (2 µl, 5 µl and 10 µl of 1 M), generating various salt particles upon 30 min incubation at 37°C. Subsequently, FBS-containing DMEM media was added to achieve final volume of 1 ml particle suspension. Absorbance at 320 nm was measured for all fabricated particles using a spectrophotometer with reference to CO3 AP.
Increment in pH, temperature and incubation time shifts the reaction equilibrium towards the forward direction by enhancing ionization and diffusion of the reactants. As shown in figure 3, particle formation was enhanced under fixed concentrations of the reactants (5 μl of 1 M BaCl2 and 2 µl of 1 M Na2SO4 , Na2SO3 , NaF, Na2CO3 or Na2HPO4 ) simply by increasing incubation period (A), pH of the HEPES buffer (B) and incubation temperature (C), as a result of the increment in energy level, therefore boosting the rate of reaction [26,27] (Figure 3).

Figure 3: (A) Effect of incubation time on particle formation. 5 µl of 1 M (a) BaCl2 was introduced into 10 μl HEPES buffered media (pH 7.5), followed by mixing of 2 µl of 1 M Na2SO4 , Na2SO3 , NaF, Na2CO3 or Na2HPO4 , generating various salt crystals upon incubation at 37°C at varying time points (0, 30 and 60 minutes). (B) Effect of pH adjustment on particle formation. 5 µl of 1 M (a) BaCl2 was introduced into 10 µl HEPES buffered media (pH ranging from 4.5-9.5), followed by mixing of 2 µl of 1 M Na2SO4 , Na2SO3 , NaF, Na2CO3 or Na2HPO4 , generating various salt crystals upon 30 minutes incubation at 37°C. (C) Effect of incubation temperature on particle formation. 5 µl of 1M (a) BaCl2 was introduced into 10 µl HEPES buffered media (pH 7.5), followed by mixing of 2 µl of 1 M Na2SO4 , Na2SO3 , NaF, Na2CO3 or Na2HPO4 , generating various salt crystals upon 30 minutes incubation at 37°C. Subsequently, FBS containing-DMEM media was added to achieve final volume of 1 ml particle suspension. Absorbance at 320 nm was measured for all fabricated particles, with reference to CO3 AP.
Higher rate of particle formation usually leads to bigger size particles [28,29]. Turbidity analysis showed that BaSO4 and BaSO3 possessed the fastest growth of particles, while BaF2 and BaCO3 were associated with the slowest growth (Figure 2B). To confirm that higher particle growth is accompanied by formation of larger particles, we estimate the average diameter for each type of the particles by Zetasizer. As shown in table 1, BaSO4 and BaSO3 were formed as particles with diameters much bigger than those of CO3 AP particles, whereas BaF2 and BaCO3 were found to be of relative small sizes. The zeta potential was estimated to be more negative for the particles formed with higher concentration (10 nM) of cation-providing salts. Images of the selected particles (BaSO3 and BaF2 ) captured by scanning electron microscope (SEM) also provide the evidence that large BaSO3 particle was the result of aggregation of smaller particles, whereas small BaF2 particle formation was accompanied by less aggregation (Figure 4 and Table 1).
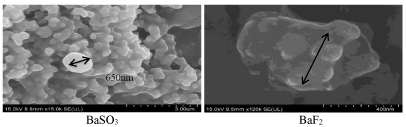
Figure 4: SEM visualization of BaSO3 and BaF2 particles. BaSO3 and BaF2 particles generated according to the protocol described in ‘Methods and materials’ section were centrifuged at 15,000 RPM for 10 seconds, followed by supernatant removal and resuspension of pelette with 1 ml milli-Q water. Fabricated salt crystals were kept on ice prior to microscopic observation. 1 µl of resuspended solution was placed onto carbon tape-coated sample holder and dried at room temperature, followed by platinum sputtering of each type of crystal samples for 60 seconds. Sputtered samples were observed at aprroximately 10-15 kV.
| Salt Formulation |
Concentration of anion-providing salt |
Size (d.nm) |
Zeta (mV) |
|
BaSO4 |
2 mM |
734 ± 41 |
-8 |
| 10 mM |
1974 ± 32 |
-10 |
|
BaSO3 |
2 mM |
506 ± 19 |
-11 |
| 10 mM |
1418 ± 33 |
-20 |
|
BaF2 |
2 mM |
218 ± 29 |
-6 |
| 10 mM |
345 ± 28 |
-15 |
|
BaCO3 |
2 mM |
243 ± 18 |
-12 |
| 10 mM |
315 ± 52 |
-16 |
|
Ba 3 (PO4)2 |
2 mM |
345 ± 61 |
-15 |
| 10 mM |
344 ± 49 |
-16 |
| CO AP3 |
- |
321 ± 51 |
-10 |
Table 1: Particle size and zeta potential of barium salt particles. Fabricated
particles were analyzed using Zetasizer to obtain average size and surface
charge of each type of crystals, with reference to CO3AP.
Results of turbidity and particle diameter show that the precipitation reactions of barium salts are driven by the type and concentration of soluble barium salts, the incubation temperature and time and finally, the pH of the reaction mixture, in order to generate a supersaturated solution, leading to particle nucleation and development into matured crystals of different sizes. Barium sulfate and barium sulfite exhibited higher number and larger sizes of particles in comparison with other salts. Particles with 20-200 nm diameter are anticipated to have excellent tumour accumulation capacity, utilizing the leaky vasculature and poor lymphatic drainage system of the tumour. Higher circulation time resulted from small particle sizes might further enhance particle buildup on tumour site [27,28]. Subsequently, particles with <100 nm in diameter is best suited for transporting genetic materials into the cell through endocytosis [30]. Negative surface charges from -6 to -20 mV were shown for various insoluble barium salts. Zeta potential, an electric potential of particle, is correlated to particle stability and surface morphology [25]. However, measured surface charge does not necessarily mean the intrinsic charge present on the particles, since the environment of the aqueous solution containing different ions could significantly influence the zeta potential. Cation-providing barium salt (BaCl2 ) in particles could create anion-binding domains, enabling the particles to bind negatively-charged nucleic acids.
Binding affinity of pDNA and siRNA towards barium salt particles
Binding affinity of DNA to barium salt crystals was assessed by labeling pGFP with PI, a fluorescence dye and forming various barium salt particles in presence of the labeled pDNA. Fluorescence images of the particles in a 24-well plate showed strong fluorescence predominantly being emitted from the aggregated particles of BaSO4 and BaSO3 as well as from the small and aggregated particles of BaF (Figure 5A). Indirect quantitative assay based on estimation of free (unbound) PI-labeled plasmid present in the supernatant (following centrifugation of various barium salt particles) (Figure 5B) indicates that BaSO4 , BaSO3 , BaF and CO3AP had significantly high binding affinity for pDNA, while BaCO3 or Ba3 (PO4 ) 2 possessed less affinity. Similar finding was observed with the same indirect quantitative assay utilizing fluorescence-labeled siRNA (Figure 5C). Lack of bright fluorescence in microscopic image of PI- plasmid-loaded CO3 AP (5-A) could be due to the existence of tiny particles emitting weak fluorescence from the relatively small no. of pDNA bound to a particle (Figure 5).
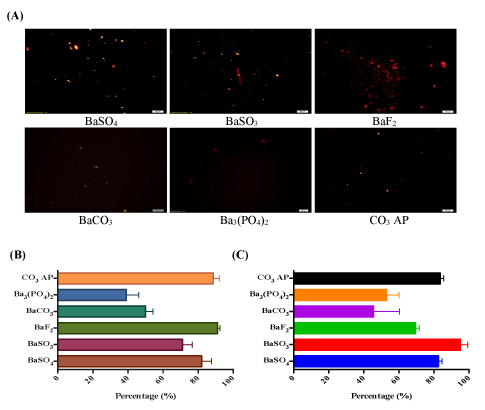
Figure 5: (A) Fluorescence microscopic observation for binding affinity of pDNA towards barium salt particles. 5 µl of 1 M (a) BaCl2 was introduced along with PI-stained pDNA (1:1 ratio) into 10 µl HEPES buffered media, followed by mixing of 2 µl of 1 M Na2SO4 , Na2SO3 , NaF, Na2CO3 or Na2HPO4 and incubation for 30 min at 37°C. FBScontaining DMEM media was added to achieve 1 ml solution. Image was captured at 10X resolution under PI filter, with reference to CO3 AP. (B) Fluorescence analysis for binding affinity of pDNA towards the particles formulated in presence of PI-labeled DNA in the same way as described above. (C) Fluorescence analysis for binding affinity of siRNA towards particles. 5 µl of 1 M (a) BaCl2 was introduced along with AF 488 siRNA into 10 µl HEPES buffered media, followed by mixing of 2 µl of 1 M Na2SO4 , Na2SO3 , NaF, Na2CO3 or Na3PO4 and incubation for 30 min at 37°C. Quantitative measurement of particles-bound pDNA/siRNA was achieved with multi-label plate reader following centrifugation of fabricated particles at 15,000 RPM for 5 minutes and aspiration of 100 µl into 96 well-plate, prior to fluorescence intensity measurement.
Cellular uptake of fluorescence-labeled nucleic acid
Cellular uptake of nucleic acid-nanoparticle complexes are usually influenced by both binding affinity of DNA to particles and average particle sizes [29]. Internalization of smaller- sized particles by cells was shown to be increased by 20-fold in comparison with larger-sized ones [31]. Fluorescence microscopic observation following 4 hr incubation of MCF-7 cells with fluorescent siRNA-loaded barium salt particles revealed significant amount of siRNA either associated with cell membranes or internalized by the cell in case of BaSO4 , BaSO3 and BaF2 particles, whereas no noticeable fluorescence signal was visualized for BaCO3 or Ba3 (PO4)2 particles (Figure 6). The very weak fluorescence signal for BaCO3 or Ba3 (PO4)2 particles could be due to their low affinity for nucleic acids, as shown in figure 5, although their sizes are relatively smaller compared to those of BaSO4 and BaSO3 particles. Despite being large in dimension, the strong signal for BaSO4 and BaSO3 particles could be explained by the notion that these particles with loaded siRNAs were mainly localized in the plasma membrane rather than being efficiently internalized through endocytosis (Figure 6).
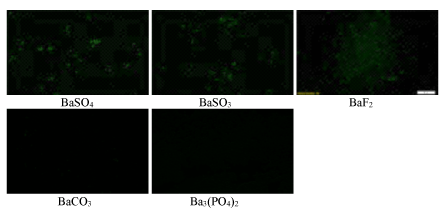
Figure 6: Fluorescence microscopic images of cellular uptake with AF 488 siRNA-loaded particles by MCF-7 cells. Different siRNA-loaded particle complexes were transferred into cells seeded at 50,000 in each well of a 24-well plate the day before the study, followed by incubation for 4 hours. Images were captured upon washing with EDTA in PBS.
Cytotoxicity profiles of barium salt particles
One of the prerequisites for clinical applications of a nanocarrier is that it should be non-toxic to the human cells. We have tested all different forms of barium salt particles in both MCF-7 and 4T1 cells at three different time points (24,48 and 72 hrs), following incubation of the particles with the cells. As shown in figures 7 and 8, all types of particles, namely BaSO4 , BaSO3 , BaF2 and BaCO3 particles except Ba3(PO4)2 ones showed similar cytotoxicity profile as that of CO3 AP, suggesting that they are relatively safe carriers. The reason behind the higher cytotoxicity of Ba3 (PO4)2 particles has yet to be understood (Figures 7 and 8).
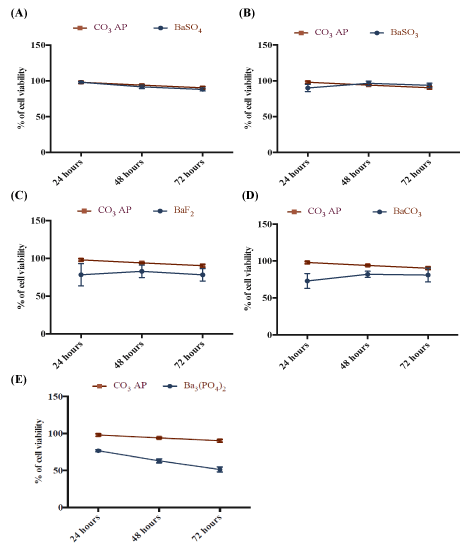
Figure 7: Cytotoxicity of barium salt particles on MCF-7 cells. 50,000 of MCF-7 cells were seeded, treated with particles and incubated for 24 to 72 hours, with media substitution following first 4 hrs of incubation. Subsequently, 50 µl of MTT was incorporated into the treated cells and medium containing MTT was aspirated after 4 hrs incubation. 300 µl DMSO was used to dissolve the formazan crystals. Spectrophotometric reading was taken at 595 nm wavelength with reference to 630 nm. Each salt type was individually compared with CO3 AP.
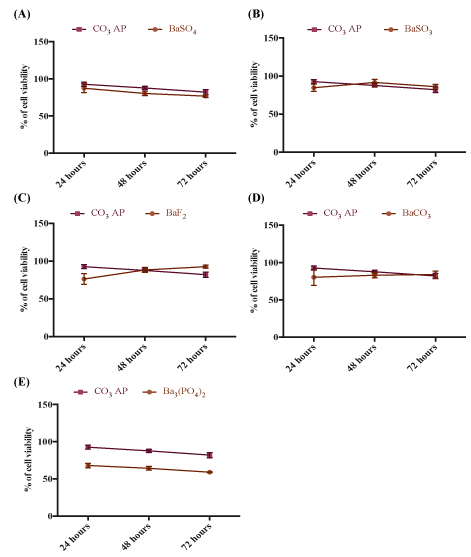
Figure 8: Cytotoxicity of barium salt particles on 4T1 cells. 50,000 of MCF-7 cells were seeded, treated with particles and incubated for 24 to 72 hours, with media substitution following first 4 hrs of incubation. Subsequently, 50 µl of MTT was incorporated into the treated cells and medium containing MTT was aspirated after 4 hrs incubation. 300 µl DMSO was used to dissolve the formazan crystals. Spectrophotometric reading was taken at 595 nm wavelength with reference to 630 nm. Each salt type was individually compared with CO3AP.
Transgene expression activity of barium salt particles
High affinity for nucleic acids with subsequent efficient cellular uptake does not gurantee that a transgene will be adequately expressed into the cell or a siRNA will be able to silence a target mRNA. For transgene expression or knockdown of an endogenous gene, a plasmid or siRNA must be released from both the particle and the endosome after endocytosis, and additionally, for a plasmid, it needs to be transported to the nucleus for expression. Reporter gene expression is an indirect, but powerful approach to confirm that a nucleic acid has been released from both the particle and the endosome, and finally transferred across the nuclear membrane. As shown in figure 9, although BaCO3 and Ba3 (PO4)2 particles did not show any activity in GFP expression in MCF-7 cells, BaSO4 , BaSO3 and BaF2 particles like CO3 AP particles dramatically led to the expression of the transgene, which could be related not only to the low affinity of DNA for BaCO3 and Ba3 (PO4)2 particles, but also probably to low acid solubility of these two particles after cellular uptake. Similar findings were noticed after transfection of both MCF-7 and 4T1 cells using luciferase plasmid (pGL3) with the help of barium salt particles (Figures 9 and 10).
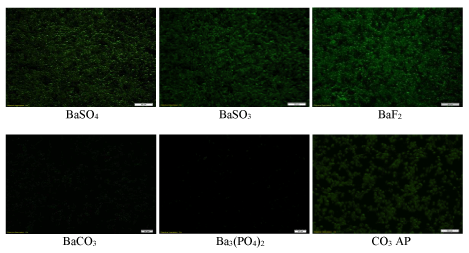
Figure 9: Fluorescence microscopic images of GFP expression after transfection of MCF-7 cells with pGFP-loaded barium salt particles. pGFP-loaded particles of BaSO4 , BaSO3 , BaF2 , BaCO3 and Ba3 (PO4)2 , along with naked pGFP and pGFP-loaded CO3 AP were used to transfect MCF-7 cells which were seeded at 50,000 in each well of a 24-well plate the day before the transfection experiment. The cells were subsequently incubated for 4 hrs and washed with 10 mM EDTA in 1X PBS, followed by 2nd incubation for a period of 48 hrs and observation under a fluorescence microscope.
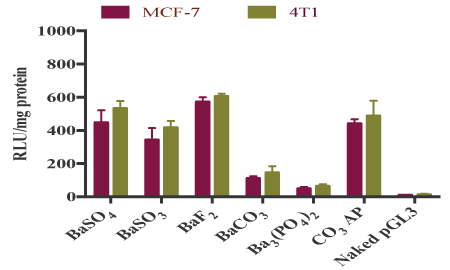
Figure 10: Transfection of MCF-7 and 4T1 cells with luciferase plasmidloaded barium salt particles. pGL3-loaded particles of BaSO4 , BaSO3 , BaF2 , BaCO3 and Ba3 (PO4)2 , along with naked pGFP and pGFP-loaded CO3 AP were used to transfect MCF-7 and 4T1 cells which were seeded at 50,000 in each well of a 24-well plate the day before the transfection experiment. The cells were subsequently incubated for 4 hrs and washed with 10 mM EDTA in 1X PBS, followed by 2nd incubation for a period of 48 hrs. The transfected cells were lysed after the removal of media, followed by centrifugation of the lysates at 15,000 RPM at 4°C for 10 minutes. 100 µl supernatant was aspirated for estimation of relative luminescence activity/mg of protein using a commercial kit and photon counting.
Intracelllular delivery of p53 plasmid and MAPK siRNA using selected barium salt particles
Based on effiacy in transgene expression level and desirable particle size, we have selected two barium salt particles, BaSO3 and BaF2 for further investigation of their potential roles in inducing cytotoxicity by facilitating intracellular delivery of p53 plasmid and MAPK siRNA. p53 is a prominent tumor suppressor with potential ability to arrest and kill cancer cells, whereas MAPK is a key enzyme of MAPK/ERK pathway responsible for ensuring proliferation of the cancer cells and therefore, silencing of MAPK expression with a specific siRNA could significantly inhibit the uncontrolled cell division. As shown in figure 11, both intracellular delivery of p53 plasmid using BaF2 and BaSO3 particles caused notable cytotoxicity in both MCF-7 (A) and 4T1 (B) cells compared with the respective particles alone. In particular, BaF2 -mediated delivery of the gene resulted in the highest toxicity in both cell lines, indicating that BaF2 nanoparticles which also enabled the highest transgene expression in two cell lines (Figure 10) would serve as a powerful tool in delivery of genebased therapeutics. On the other hand, BaSO3 particles were found to be more efficient than BaF2 in causing more cell deaths in both MCF-7 (C) and 4T1 (D) cells through intracellular delivery of MAPK siRNA, suggesting that BaSO3 particles and MAPK siRNA might synergistically contribute to the significant cytotoxicity. Intracellular delivery and expression of p53 gene is more challenging than delivery and silencing activity of MAPK siRNA due to the fact that silencing of a target mRNA happens in cytosol where as transgene expression requires nuclear translocation of a plasmid DNA to initiate transcription in nucleus and subsequently, translation in cytoplasm. Barium salt particles like other types of delivery vectors, e.g., CO3 AP could bind to the anionic cell membrane through ionic interactions using their cationic salt components (Ba2+), resulting in endocytosis and subsequent degradation at low endosomal acidic pH. As a consequence, DNA or siRNA could be released and subjected to endosomal escape according to ‘proton-sponge’ hypothesis [10,14] (Figure11).
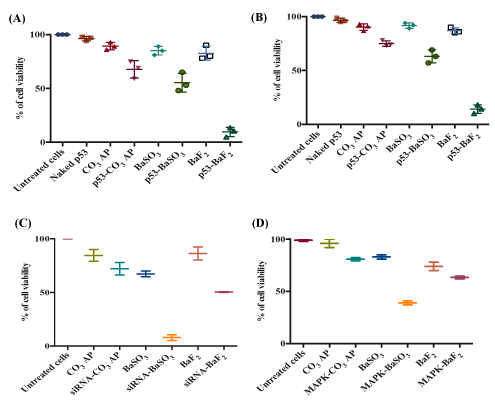
Figure 11: Cytotoxicity outcome following introduction of p53 plasmid with BaSO3 and BaF2 particles. Cell viability of (A) MCF-7 and (B) 4T1 cells after treatment with p53-loaded barium salt particle complexes. 5 mM BaCl2 was mixed with 500 µg of p53 prior to incorporation of 2 mM Na2SO3 or NaF in 10 µl of HEPES solution. Cell viability of (C) MCF-7 and (D) 4T1 cells after treatment with MAPK siRNA-loaded barium salt particle complexes. 5 mM BaCl2 was mixed with 10 nM MAPK siRNAs prior to incorporation of 2 mM Na2SO3 or NaF in 10 µl of HEPES solution. Each set of experiments included untreated cells, CO3 AP and unloaded salt particles as controls. Nucleic acid-loaded particles were transferred into each well of a 24-well plate containing breast cancer cells seeded at 50,000 cells/well on the day before. The cells were incubated for 48 hrs after replacement of particle-containing media with serum-supplemented media following first 4 hr incubation in presence of the various particles and subsequent treatment with 5 mM EDTA in PBS. Finally, 50 µl of MTT was incorporated into the treated cells, and the media containing MTT was aspirated after 4 hr incubation. 300 µl DMSO was added to dissolve the formazan crystals. Spectrophotometric reading of viable cells was taken at 595 nm wavelength with reference to 630 nm.
Analysis of activation of MAPK and AKT pathways after intracellular delivery of MAPK siRNA with selected barium salt particles
Since MAPK and AKT pathways are known to play key roles in cancer cell proliferation and survival, respectively, and both of the pathways can cross-talk to each other, we have investigated the activation levels of the two pathways following intracellular delivery of MAPK siRNA with the help of BaSO4 , BaSO3 and BaF2 particles. As shown in figure 12, BaF2 particles enabled knockdown of MAPK and AKT, leading to inhibition of their phosphorylation to a remarkable extent in MCF-7 cells, which is similar to the silencing outcome of CO3 AP particles. The high capacity of BaF2 particle in siRNA-mediated knockdown was correlated with its small diameter, high affinity for nucleic acids, effective cellular uptake and p53- and MAPK siRNA- mediated enhanced cytotoxicity. The relatively lower knockdown efficacy of BaSO4 compared to that of BaSO3 was probably due to the larger size diameter of the former, thus hindering the cellular uptake of the siRNA. The overall lower knockdown activity in 4T1 cells might be due to faster growth of the cells with the result of relatively lower uptake of the siRNA-loaded particles by the cells (Figure 12).
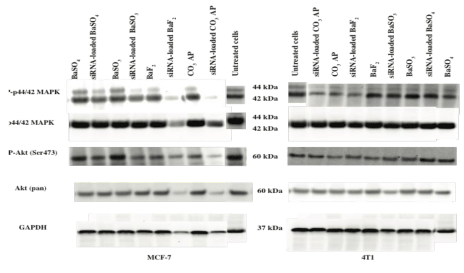
Figure 12: MAPK protein expressions following treatment of MCF-7 and 4T1 cells with MAPK siRNA-loaded selected barium salt particles. Proteins obtained from lysates of the treated cells were run on SDSPAGE and transferred to PVDF membrane, followed by incubation with primary antibodies raised in rabbit against phospho-p44/42 MAPK, p42/42 MAPK, phospho-Akt (Ser473) and Akt (pan). HRPconjugated goat anti-rabbit secondary antibody was used to detect the chemiluminescence signals. Predicted bands for pMAPK, TMAPK, pAkt and TAkt are at 44, 42 and 60 kDa, respectively. GAPDH was used as loading marker with bands achieved at 37 kDa.
Conclusions
Among the barium salt particles, BaF2 is the most potential nanocarrier to deliver plasmid DNA and siRNA into breast cancer cells, partially owing to its small size, strong binding affinity for nucleic acids, efficient cellular entry and capability to release the nucleic acids from the particles and the endosomes, thus enabling either silencing of a target mRNA with a specific siRNA or translocating pDNA into the nucleus for subsequent transcription. Both BaF2 and BaSO3 particles demonstrated high potential in delivering p53 gene and MAPK siRNA for inducing death in breast cancer cells. To our knowledge, this is the first report on efficient intracellular delivery of nucleic acids with the help of BaF2 and BaSO3 nanoparticles. Manipulation of the barium salt formulations with addition of ligands might improve their efficacy for nucleic acid delivery. A pre-clinical study based on an animal model of breast cancer would pave the way to further explore the potential of barium salt particles for intravenous delivery of nucleic acid-based therapeutics.













