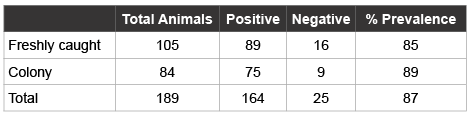
Table 1: Seroprevalence of HVP-2 antibodies comparing exposure rates in freshly-caught and captive colony baboons
Sharon Chepkwony1,2 Nicholas M Kiulia1 Ruth Nyakundi3 Michael Gicheru2 Atunga Nyachieo1*
1Molecular Biology Unit, Department of Reproductive Health and Biology, Institute of Primate Research, Karen Nairobi Kenya*Corresponding author: Dr. Atunga Nyachieo, Molecular Biology Unit, Department of Reproductive health and Biology, Institute of Primate Research, P.O Box 244481-00502, Karen Nairobi, E-mail: anyachieo@yahoo.com
Background: Herpes simplex virus types 1 and 2 (HSV-1 and HSV-2) are prevalent in humans and cause significant morbidity and even mortality. There is no cure for HSV, and available drugs only shorten the recovery period and may lengthen time between recurrences. Development of an animal model that closely resembles humans is crucial for testing of new treatment interventions. Baboons carry a virus (Herpesvirus papio 2; HVP2) that is closely related to HSV. HVP2 produces infections in baboons that are clinically similar to those caused by HSV in humans. While baboons show promise as an animal model for studying HSV, the overall and comparative prevalence of HVP2 in male and female wild baboons is not known.
Methods: In this study, the sero prevalence of HVP2 in baboons was determined by detection ofanti-HVP2 antibodies in sera from 189 wild baboons captured from different regions of Kenya. A PCR test with specific primers targeting the UL41gene of HVP2was used to confirm the presence of HVP2.
Results: In total, 87% of the baboons had been exposed to HVP2. About 90% of female and 83% of male baboons were sero positive for HVP2. There was no significant difference in the prevalence of HVP2 in baboons from different geographic regions. PCR test confirmed the presence of HVP2 strain in a sero positive baboon captured from Laikipia County of Kenya.
Conclusion: The prevalence of HVP2 was high in baboons from different regions within Kenya. There was no significant difference in HVP2 infection rates in females and males. Information on sero prevalence and molecular epidemiology of HVP2 will positively influence the use of baboons as models to study the pathogenesis of HSV and test vaccine strategies.
Herpes simplex virus (HSV) is an alpha-herpes virus that infects humans. There are two serotypes: HSV-1 and HSV-2. HSV-1 primarily causes oral infections while HSV-2 is the leading cause of genital herpetic infections. HSV-2 is a co-factor in HIV-1 acquisition and transmission as well as a cause of neonatal herpes infections [1,2]. HSV establishes latent infections in sensory neurons, and the virus can periodically reactivate from latency, resulting in recurrent infections. Primary genital HSV-2 infections that are acquired late in pregnancy are estimated to cause about 80% of neonatal herpes infections [3-6], and can result in significant morbidity to the newborn child [7,5]. Recurrent HSV-2 infections are estimated to cause less than 5% of neonatal herpes transmission [6,4], while HSV-1 is responsible for about 20% of neonatal herpetic infections [6,4]. The acquisition of genital herpes during pregnancy has been associated with spontaneous abortion, prematurity, and congenital and neonatal herpes [8-10]. Active HSV infection (shedding of virus) at the time of delivery may result in infection of the newborn infant during passage through the birth canal or by ascending infection after rupture of the membranes. Neonatal herpes is a serious condition with about 60% mortality [6]. HSV has no cure and attempts to develop vaccines and drugs to cure the disease have been futile.
Infection with HSV-1 occurs worldwide, equally between the sexes. There are no seasonal variations for the infection with this particular virus. In the United States, it is estimated that there are approximately 500,000 primary infections annually [11]. HSV-2 is a common infection in many countries, with prevalence in some regions, such as sub-Saharan Africa, higher than in the USA [12]. According to the U.S. Centers for Disease Control and Prevention, 45 million people in the United States aged 12 years and older, or 1 out of 5 of the total adolescent and adult population, are infected with HSV-2. Most data from Central and South America are from women, in whom HSV-2 prevalence ranges from about 20% to 40%. Prevalence in the general population in developing Asian countries appears to be lower (10-30%). Studies in the African continent have shown variation in HSV-2 prevalence across diverse populations of women, ranging from 22% among adults in Tanzania and 68% among adults in Kenya to 90% among sexual workers in Zaire [12,13]. In western Kenya, an HIV survey among women aged 13-34 years of age conducted in 2003-4 revealed an HSV-2 prevalence of 53% [14]. A more recent National AIDS Indicator Survey among Kenyan women aged 16-64 years estimated HSV-2 prevalence to be 42% [15].
Viruses related to HSV that are endemic in various non-human primates (NHPs) such as chimpanzees, gorillas, orangutans, baboons, cynomologus and rhesus macaques, and marmosets are capable of infecting humans and causing diseases that are sometimes fatal [16].Since they are phylogenetically very closely related to humans, NHPs are the best models for investigating the biological roots of human diseases [17]. After initial interspecies transmission, these viruses have the potential to evolve and disseminate in the human population. Understanding of the initial steps of emergence of zoonotic infections and associated diseases remains poor. NHPs share ~98% of human genes. For this reason, they play a critical role to biomedical research in the advancement of human and animal health by targeting the cause, progression, prevention and treatment of a wide variety of diseases.
There are several neurotropic alpha-herpesviruses that are related to HSV that infect NHPs [18,19]. All these viruses produce diseases in their natural hosts that are similar to HSV in humans. Macacineherpesvirus 1 (Monkey B virus; BV) is prevalent in macaques (Macaca spp.). When transmitted to humans via bites or scratches, BV replicates in the peripheral epithelial tissue where it invades the sensory ganglia. The virus then progresses into the central nervous system (CNS) where it continues to replicate and spread resulting in extensive tissue damage leading to death in most cases [16]. Cercopithecineherpesvirus 2 or Simian Agent 8 (SA8) infects African green monkeys (Cercopithecusaethiops). Herpesvirus papio2 (HVP2) is very closely related to SA8 and is indigenous in baboons (Papio spp.) Herpesvirus saimiri 1and Herpesvirus ateles 1 are common in squirrel monkeys and spider monkeys, respectively. Of particular relevance to this study,baboons are closer in terms of phylogenetic relationship to man than the other mentioned NHP species [20].
As with other alpha herpesviruses, HVP2 causes oral, urogenital and neuronal infections that closely resemble those caused by HSV in humans [21]. Infected adult baboons develop genital lesions whereas oral lesions represent the predominant form of disease in younger animals [22]. The lesions range from vesicles to ulcers. Spontaneous reactivation from latency has been demonstrated, a pattern consistent with HSV infection of humans [21]. Acquisition of genital infections is primarily associated with the onset of sexual activity in baboons.Secondary bacterial infection is common, and epidermal necrosis with central erosions and peripheral parakeratosis is also observed [23]. The prevalence of HVP2 was determined in several groups of captive and wild-caught baboons adult baboons. Although the number of animals tested was relatively small, over 90% of wild-caught adult olive baboons (Papioanubis) from Kenya and chacma baboons (P. ursinus) from South Africa were found to have antiHVP2 antibodies. Similarly, approximately 85% of captive breeding colony baboons (P. anubis and P. cynocephalus) were seropositive for HVP2 [24]. There is however no available data on HVP2 prevalence in baboons from different geographical regions of Kenya. This study therefore determined HVP2 exposure rates and identified one circulating strain in baboons from selected geographical regions of Kenya.
Baboons (P. anubis) were trapped in Laikipia, Nyandarua, Kirinyaga, Machakos, Kajiado and Nairobi counties of Kenya, transferred to holding cages, and transported to the Institute of Primate Research (IPR) quarantine area. At IPR, baboons were housed in special facilities in accordance with guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC). The baboons brought to the Institute were used for many different research studies.
Two weeks after arrival at IPR, wild-caught baboons were anesthetized with a 3:7 mixture of 2% xylazine and 10% ketamine hydrochloride (0.1 mg/kg) for physical examination and processing [25]. Baboons were sampled (blood for serum and swabs for virus isolation) at this time. Venous blood was collected from 105 baboons. Blood was processed and serum stored at -20°C. Eighty four additional serum samples were acquired from the IPR serum bank. These sera were from wild-caught baboons that had been housed at IPR for more than three months. Oral swabs were collected from 31 juveniles while genital swabs were collected from 69 adult baboons. Sterile cotton-tipped applicators were used as swabs and placed in 1ml transport medium (Digene® Female Swab Specimen Collection Kit, QIAGEN).
Antigens for ELISA (HVP2 infected cell and uninfected cells) were generously provided by Dr. R Eberle of Oklahoma States University Center for Veterinary Health Sciences, USA [26]. Lyophilized antigens were resuspended in sterile water (to reconstitute to 0.5% Triton X-100 and 0.01% sodium dodecyl sulphate) and extracts clarified by centrifugation at 14,000 rpm for 30 sec. Supernatants (antigen) were aliquoted and stored at -80⁰C.ELISAs were run basically as described [26] except that peroxidaseconjugated anti-human IgG was used for detection of baboon IgG. TMB was used as substrate and OD read at 630nm.
Viral DNA was extracted from swabs of seropositive animals using the ZYMO RESEARCH ZR -DNA Extraction Minikit (Inqaba, South Africa) according to the manufacturer’s protocol.DNA was stored at -20⁰Cuntil used for PCR.Two primers were designed to amplify a 500bp region of the UL41 (virion host shut-off protein) gene of HVP2 (TK1,5’TTCACCGTGGGCTGGACTGG3’ and TK2,5’GCGGTTCTGGAGCTCGGACCA3’). The primers were designed based on aligned sequences of 7 HVP 2 strains of highly conserved regions. PCR was performed using a PCR master mix (AZymogen). PCR was run using an initial denaturation step (94°C for 5 min) followed by 30 cycles of denaturation at 94°C for 60 sec, annealing at 60°C for 60 sec, and elongation at 72°C for 120 sec. A final extension of products was carried out at 72°C for 4 min.
Pearson Chi- χ2 square (two by two table) was used to compare the number of positive samples to negative samples (proportions) for both female and male baboons in captive vs. wild caught animals. Graph Pad Prism was used for data analysis [27]. Percentages of exposed animals were determined where age, sex and location were compared. A P value of <0.05 was considered significant.
A total of 189 baboons from six counties in Kenya including baboons both newly caught and those that had been in the facility for over 3 months were sampled. A comparison in HVP2 exposure frequencies in freshly caught versus captive IPR colony baboons was made (Table 1). The prevalence of HVP2 in captive colony baboons was only slightly higher than in freshly caught wild baboons and the difference was not significant(X2 =0.83; df=1; P>0.05). Since the difference in HVP2 positivity between IPR colony and wild baboons was not significant, animals were treated as a single group for further analyses.

Table 1: Seroprevalence of HVP-2 antibodies comparing exposure rates in freshly-caught and captive colony baboons
The prevalence of HVP2 was also compared among baboons from different regions (Figure 1), by gender, and by age (Tables 2 and 3 respectively). The counties (number of baboons) sampled were Laikipia (65), Nyandarua (33), Kirinyaga (31), Machakos (24), Kajiado (16) and Nairobi (20). These counties experience different climatic conditions, with Nyandarua County being the coldest and Machakos and Kajiadothe hottest. Findings from this study showed a high prevalence of HVP2 infection in baboons from all the regions ranging from 71% in Kirinyaga county to 100% in Machakos, Kajiado and Nairobi counties.
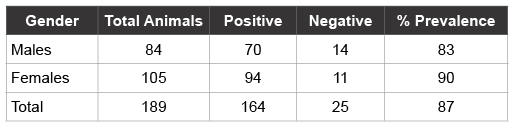
Table 2: Seroprevalence of HVP2 antibodies comparing gender exposure rates
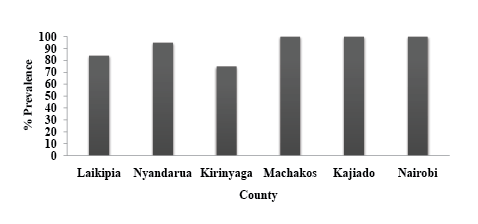
Figure 1:Percentage prevalence of HVP2 in baboons from selected regions of Kenya seroprevalence was determined by ELISA
To assess the age distribution of HVP2 seropositivity in wild baboons, animals were divided into three age groups: Juveniles (1-3yrs or months), Sub-Adults (4-6yrs) and Adults (>7yrs). Significantly more adults and subadults were found to be seropositive as compared to juveniles (Table 1; X2 =40; df=2; P<0.05).
To confirm the presence of HVP2 in seropositive baboons, PCR was used to amplify a 500bp region of the HVP2 UL41 gene. Out of 100 baboons swabbed, 76 were seropositive for HVP2. DNA from oral and genital swabs of these 76 animals were tested. Thirty two of these amplified DNA samples showed visible bands on agarose gel. Forty two percent of animals that were positive by ELISA were also positive by PCR (Figure 2).
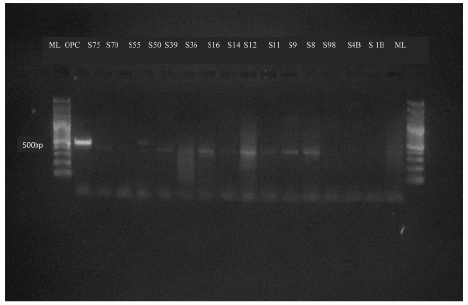
Figure 2: PCR detection of HVP2 in wild baboons. PCR amplified a 500bp region of the virion host shut-off UL41gene. ML, molecular size ladder; OPC, positive control; Sn, sample number, where n is the baboon number
One of the PCR products was sequenced. Results showed 98.0-98.7 identity to 7 different HVP2 strains, being slightly higher against mousedefined neurovirulent HVP2 strains (98.2-98.7%) than to a pathogenic strains (98.0%) [28]. Sequence identity to the most closely related virus SA8 was only 94.0%.
The current study provides strong evidence of natural exposure of wild olive baboons (Papioanubis) from selected geographical regions in Kenya to HVP2. It also demonstrates a high prevalence of HVP2in these wild baboons.
The regions in which the baboons were captured experience different climatic conditions. Nyandarua county and particularly Aberdares are part of the Kenyan highlands that experience the coldest weather conditions. Machakos and Kajiado counties on the other hand, are the hottest of the six counties from which the baboons were sampled. Despite different climatic conditions, the prevalence of HVP2 was equally high in all regions, with over 70% of animals from each region having been exposed. This likely indicates that climatic differences have no influence on the infection rates of HVP2 in baboons. However, baboons are known to migrate from one region to another. The prevalence rates we found in captive baboons are similar to those reported by other investigators in wild and captive colony baboons [29,24,22]. In any case, the prevalence of HVP2 infection in baboons is universal.
The prevalence of HVP2 antibodies was found to be high in adult baboons (94%) and sub- adults (90%) as compared to juveniles (52%). Again, similar results were obtained regarding HVP2 transmission in a captive baboon colony [29]. The age difference in prevalence can be explained by the fact that adult baboons are sexually active while most sub-adults are at the onset of sexual activity, so both oral and sexual transmission of HVP2 can occur. Being sexually immature, juveniles likely acquire HVP2 as an oral infection. A study investigating shedding and transmission of HVP 2 in captive baboons [23] documented both of these modes of HVP2 transmission. The virus that was isolated from a mother who was shedding the virus was indistinguishable from the virus isolated from the oropharynx of her infant and several other co-housed infants. Thus, transmission of HVP2 appears to be similar in captive and wild baboons.
The rate of HVP2 infection was slightly higher in females (90%) as compared to males (83%), although this difference was not statistically significant. This is consistent with a study that was done in human population comparing HSV-2 infection rates between men and women [30]. Biologically, females are more vulnerable to sexually transmitted infections as compared to males. During heterosexual sex, the exposed surface area of the vagina and labia is larger in females than the vulnerable surface area in males [31] and these increases chances of transmission in females. Thus, it is not surprising that HVP2 prevalence is somewhat higher in female vs. male baboons.
There was no significant difference in HVP2 prevalence in IPR captive colony baboons that had been in the facility for more than 3 months (85%) and the freshly caught wild baboons (89%). This can be attributed to the fact that baboons live in troops in the wild. Troop sizes vary between 5 and 250 animals (usually ~50) depending on specific circumstances, especially species and time of year [32]. Housing conditions permitting, the same behavior is observed in captive baboons. Baboons readily breed in captivity, so HVP2 can easily be transmitted to other baboons that are sexually mature in captivity or in the wild. This explains why there is a small difference in HVP2 infection prevalence between the captive colony baboons and the wild-caught baboons.
Fewer animals were positive by PCR as compared to those that were seropositive by ELISA (42% of ELISA positive baboons). These results can be attributed to the fact that HVP2 is usually latent in sensory ganglia and infectious virus can only be detected when the animal is shedding. Thus, virus would only be present in swabs during reactivation. Shedding of HVP2 is not frequent in captive baboons [23]. These investigators found that out of 342 swabs collected from 128 animals over 1.5years, infectious virus was detected in only 13 swabs (3.8 %). The study also showed that shedding of the virus was higher among infants as compared to adults and most shedding was in the oral cavity. Most of the swabs collected during this current study were from the genital region of the adult baboons. A higher incidence of genital shedding (42% of the samples) was reported in this study, indicating that the stress of capture and transport possibly reactivates the virus. This has also been observed for reactivation of monkey B virus in macaques [33].
Many strains of HVP2 have been isolated and sequenced [34,28]. The strain identified in this study showed 6-9 nucleotide differences from sequenced strains, and some of these substitutions were consistent against all sequenced isolates. All HVP2 isolates sequenced to date were isolated from baboons in the USA, so it appears that HVP2 found in wild Kenyan baboons may be slightly different from US HVP2 isolates. It was interesting that the sequence determined in this study was slightly more identical to mouse-defined neurovirulent HVP2 isolates than to a pathogenic HVP2 isolates the “neurovirulent strains” rapidly invade the CNS in mice causing death, but produce self-limiting lesions similar to those caused by “apathogenic strains” when inoculated into baboons [35]. This raises concerns about the zoonotic potential of HVP2 found in wild baboons given that humans are phylogenetically close to baboons. While no cases of human infection with HVP2 have been reported, there is nothing to suggest that humans cannot be infected with the virus.
The seroprevalence of Herpesvirus papio 2 infection in baboons in Kenya is 87%. The prevalence was high in baboons from different regions within Kenya. There was no significant difference in HVP2 infection rates in females and males. Infection rates of HVP2 are higher in adult and sub-adult baboons as compared to the juveniles, suggesting that HVP2 infection and transmission is associated with the onset of sexual activities in olive baboons (Papioanubis). Results regarding HVP2 prevalence in wild Kenyan baboons were very similar to that reported for captive baboons maintained in troops.
We acknowledge Dr. R. Eberle from the Department of Veterinary Pathobiology, Center for Veterinary Health Sciences, Oklahoma State University, Still water Oklahoma, USA for proving us with Antigens, primers and controls used in this study. We also acknowledge technical support prvided by Erick Omolo and Mary Galo, IPR.
Download Provisional PDF Here
Article Type: Research Article
Citation: Chepkwony S, Kiulia NM, Nyakundi R, Gicheru M, Nyachieo A (2016) Sero-Prevalence of Herpesvirus Papio 2 in Wild-Caught Olive Baboons from Selected Regions in Kenya. J Emerg Dis Virol 2(3): doi http://dx.doi.org/10.16966/2473-1846.118
Copyright: © 2016 Chepkwony S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Publication history:
All Sci Forschen Journals are Open Access