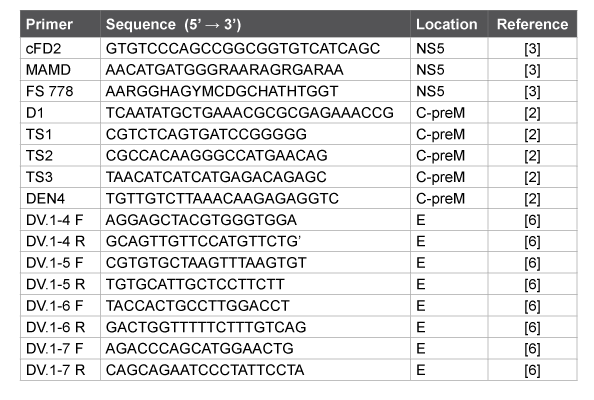
Table 1: List of primers used for PCR and sequencing
Molecular Typing of Imported Dengue Virus (DENV) Isolates from Patients in the Far East of Russia
Pukhovskaya NM1* Morozova OV2,3 Bakhmetyeva SV1 Zdanovskaya NI1 Belozerova NB1 Pochinkova NA1 Ivanov LI1
1Khabarovsk antiplague station Rospotrebnadzor, 7 Sanitarny Bystreet, 680037, Khabarovsk, Russia*Corresponding author: Natalia M. Pukhovskaya, Khabarovsk Atiplague Station Rospotrebnadzor, 7 Sanitarny Bystreet, 680037, Khabarovsk, Russia; Tel: +7 4212334597; Fax: +74212334526; E-mail: pukhovskaya@mail.ru
Background: Dengue remains the most rapidly spreading mosquito-borne viral infection in the world with 30-fold growth during the last 50 years and its expansion to new areas. Dengue virus transmissions are effected by urbanization and international travel. Detection, identification and quantitation of Dengue virus beyond its endemic regions during global warming are necessary to estimate an epidemiological risk.
Methods: Dengue virus strains were isolated from blood samples of patients with unknown fever. To confirm clinical diagnosis both RTPCR and ELISA to detect IgM/IgG were used. Sequencing of RT-PCR products corresponding to C-prM, E and NS5 genes with subsequent phylogenetic analysis performed for molecular typing.
Results: During 2012-14 Dengue fever imported cases were confirmed in blood samples of 13 from 95 patients with unknown fever (13,7%) in Khabarovsk, Primorye and Sakhalin regions of the Far East, Russia. Using permissive tissue culture Vero E6 and susceptible suckling mice five Dengue strains of 1 and 3 types have been isolated. RT-PCR with Dengue serotype-specific primers and phylogenetic analysis of nucleotide sequences of RT-PCR products corresponding to C-prM, E and NS5 genes revealed Dengue viruses 1, 2 and 3 types.
Conclusions: For the first time in Russia Dengue fever imported cases were confirmed by means of RT-PCR, ELISA and virus isolation. Molecular typing revealed Dengue viruses 1, 2 and 3 types in blood sample of patients after their return from travel to subtropical endemic regions.
Molecular typing; Dengue virus; RT-PCR
Dengue virus (DENV) remains an emerging and major public health concern. High prevalence, with at least 50 million new cases each year [1], wide geographic distribution with approximately 2.5 billion people living in dengue endemic regions, co-circulation of multiple serotypes (hyperendemnicity), neurovirulence and hyperimmune responses are severe risk factors. Outbreaks of Dengue have been documented since 1799 in Egypt [1]. Though sporadic prior to World War II, the frequencies of Dengue outbreaks increased significantly after the war and were accompanied by the growth of severe Dengue rate compared to Dengue fever according to the World Health Organization (WHO) classification [1 and references therein]. Currently, it has become the most rapidly spreading mosquito-borne viral infection in the world and during the last 50 years its incidence has risen 30-fold with geographic expansion to new areas. Currently, the DENV breached the subtropical temperature barrier of the previously known endemic regions of the Americas and the Pacific, extended its reach as far north as Nepal, China and France and as far south as Argentina and expanded from urban to rural settings [1-11]. Incidences in South East Asia and the Western Pacific account for greater than 75% of the global burden of Dengue.
Four genetically and antigenically distinct serotypes (DENV-1 to DENV-4) of the flavivirus are transmitted among humans and higher primates by mosquitoes of the genus Aedes. Humans are the main amplifying hosts of the virus in the urban cycle. The increased intensity of DENV transmission is effected by the rise in number and size of densely populated urban cities and by international travel which results in the continuous importation and exchange of Dengue serotypes and genotypes. The epidemiology of Dengue is affected by a complex interplay of DENV virulence, transmissibility, host susceptibility and immune response, environmental and climate factors, hyperendemnicity and epidemic activity [1-8].
DENV possesses a positive-sense, single-stranded RNA genome, which is approximately 10,700 bases in length and contains a single open reading frame with the following gene order 5’-C-prM/M-E-NS1-NS2ANS2B-NS3-NS4A-NS4B-NS5-3’. Like a majority of RNA viruses, DENV commonly exists as a swarm of viruses with varying genomes known as quasispecies. The variations are predominantly the result of the lack of proof-reading capability in the virally-encoded RNA-dependent RNA polymerase, the production of large numbers of viral genomes and the absence of RNA repair systems in both host eukaryotic cells and in the virus causing a high average mutation rate of 10-3 to 10-5 substitutions per nucleotide and per round of replication [12,13].
In Russia, during the last years, a few Dengue fever cases have been confirmed by means of ELISA to detect DENV-specific antibodies [9-11]. In the European part of Russia Dengue clinical cases were registrated after travels of patients to Indonesia (22), Thailand (11), India (3), Vietnam (3), Venezuela (2), Singapore (1), Sri Lanka (1), Malaysia (1), Costa Rica (1) and Dominican Republic (1) [9], whereas in the Far East Russia the main part of Dengue fever were after visits to South East Asia, mainly Thailand [10,11].
Dengue is not particularly associated with neurovirulence. Severe Dengue is associated with plasma leakage, bleeding and shock. A less common manifestation is hepatitis. Early confirmed diagnosis may be valuable for fulminant infection with possible neurological complications and life-saving. Diagnostic methods include DENV isolation, as well as detection of viral RNA, antigens and/or antibodies used singly or in combination. Among them the most sensitive methods are virus isolation and reverse transcription with PCR that result in a confirmed diagnosis of an acute infection if the specimens (whole blood, serum, plasma, tissue) are taken within 1-5 days after onset of symptoms. ELISA detection of both IgM and IgG antibodies in a single serum without an additional clinical sampling may suggest a probable diagnosis but additional confirmation is needed [1-6,9-11,14].
Our research was aimed at detecting and identifying an etiological agent of an acute unknown fever among patients in the Far East of Russia.
A total of 183 blood samples from 95 patients with unknown fever were kindly provided by the staff of hospitals in Khabarovsk, Vladivostok, Yuzno-Sakhalinsk and Blagoveschensk (the Far Eastern part of Russia) during 2012-2014.
DENV strains were isolated from patient blood samples and human lymphocytes by means of infection of Vero E6 tissue culture or both intracerebral and subcutaneous infection of 1-2 days old ICR mice [4]. Cytopathic effect was observed in 6-8 days after original infection or after 4-5 days postinfection for the subsequent passages. In 10-13 days postinfection of newborn suckling mice the following disturbances were revealed: languor, tremor, paresis and growth retardation. The DENV RNA was detected in the infected tissue cultures and mouse brain suspensions by means of RT-PCR [2,3,6].
Elisa was used for the detection of IgM/IgG against DENV and specific antibodies against West Nile virus (WNV) with the kits “Euroimmun” (Germany) and “Bioservice Biotechnology Company Ltd” (Moscow, Russia) as well as with the test systems kindly provided by Doctor Victor F. Larichev (Ivanovsky Institute of Virology of the Russian Ministry of Health, Moscow, Russia [9]). Detection of IgM/IgG antibodies against the tick-borne encephalitis virus (TBEV) was done using the kit “Vector Best” (Novosibisk, Russia) according to the manufacturer’s instruction.
Detection of DENV NS1 antigen was performed by using a rapid immunochromatopraphic test in human serum, plasma or whole blood (Standart Diagnostics, Korea; MT Promed Consulting GmbH, Germany).
Total nucleic acids were isolated using “Riboprep” kit (“InterLabService”, Moscow, Russia). Reverse transcription with random N6 primer was carried out with “Reverta L” kit (“InterLabService”, Moscow, Russia) according to the manufacturer’s instructions. PCR with primers specific to DENV C-preM and E genes and flavivirus-specific primers corresponding to NS5 gene (Table 1) and synthesized in “Syntol” (Moscow, Russia) was carried out using the kit «AmpliSensPCR» (“InterLabService”, Moscow, Russia). The WNV RNA was detected by using «AmpliSens PCR WNV – FL» (“InterLabService”, Moscow, Russia); later in 2014 «AmpliSens Dengue virus type – FL» (“InterLabService”, Moscow, Russia) was used for detection and molecular typing of DENV RNA according to the manufacturer’s instructions.

Table 1: List of primers used for PCR and sequencing
Nucleotide sequences of RT-PCR products were determined using BigDye 3.1 Terminator Cycle Sequencing Kit and DNA analyzer ABI 3500 (Applied Biosystems, USA). Phylogenetic analysis was performed using MEGA 6.06 [15].
GenBank (http://www.ncbi.nlm.nih.gov) accession numbers of the DENV isolates determined in our study are KJ396963-KJ396967, KJ417840- KJ417844; for reference strains used for phylogenetic analysis - HG316481; HG316482; KJ396964; U87411; NC 001474; HG316483; HG316484; KJ396967; KJ417840; KJ417841; KJ417843; KJ417844; JN575594; JQ993305; JQ993306; FJ196844; AB608787; KC172831; JN054256; KF954949; GU370053; DQ18180; KJ734727; JF262782; AY762085.
During 2012-2013 essential growth rate of Dengue fever throughout the world and particularly in the South-East Asia was observed [1,5-11]. In Thailand periodic fluctuations of DENV annual infection rate varied from 8,100 to 174,285 according to the data from Thailand national annual Dengue incidence surveillance [7,8]. All DENV serotypes are known to persist simultaneously in Thailand, with one of them emerging as a cause of periodic moderate-to-severe epidemics [5]. During 2012- 13 the disease rate was there extremely high: 74,250 patients with 79 deaths in 2012 [7]; 152,768 patients with 132 deaths in 2013 [8]. Severe epidemiological situation is emerging especially in continental Thailand with world resorts, recreation zones and numerous travelers evidently influenced number of DENV imported cases in surrounding countries of even temperate climate [1,5-11].
Among patients with DENV infection in Russia the prevalent clinical manifestation was a febrile illness with frequent headache. The single case of rush and myalgia was registrated. It was associated with the detection of the DENV-specific IgM with titers exceeding those against WNV and TBEV at least in 4 fold; IgG antibodies; the DENV RNA and NS1 antigen as well as the isolation of the DENV in tissue cultures or in newborn mice (Table 2).
Patients with fever of unknown etiology were from the Khabarovsk region (68), Vladivostok and other towns of Prymorie (23) as well as from the island Sakhalin (2) and Amur region (2). Among 85 persons (the place of infection of whom was known), 63 (74,1%) patients previously visited Thailand; 10 – Vietnam, 3 – the Philippines, 2 – Bali, 2 – Guam, 1 – Chile, 1- India, and 3 – Africa (Egypt, Mali and Zambia). RT-PCR of 183 blood samples including grume, lymphocytes and sera from 95 patients with tropical fever allowed us to detect flavivirus and/or DENV RNA in the blood of 16 patients (16.8%); whereas WNV RNA was revealed in none of them. Among 16 PCR positive samples, 11 were collected during the first week of clinical manifestations (5 of them with the DENV-specific IgM antibodies); 4 samples - during the 2nd and 3d weeks (contained both viral RNA and specific antibodies); 1 sample - during the 4th week of clinical manifestations, contained neither antibodies. One should note that DENV strains of 1 and 3 types can be isolated from PCR negative samples (2 strains 2759 and 2996) as well as from human blood containing IgM antibodies (Table 2). Molecular typing by means of PCR with DENVspecific and universal flavivirus primers with subsequent sequencing of RT-PCR products revealed 5 isolates DENV-1; 3 – DENV-2, 5 – DENV-3; twelve of them were from patients after their travel to Thailand and one – after visiting Vietnam (Table 2). Thus, Dengue infection was confirmed for 13 of 95 patients with unknown fever (13.7%) by means of PCR and sequencing. In blood samples from 5 patients, flavivirus RNA and DNVspecific IgM antibodies allowed us to suspect Dengue fever.
DENV RNA was detected in both blood serum and cells using RT-real time PCR with threshold cycles (Ct ) varying in a range 11,46 – 21,95 that corresponded to 2,7 × 105 - 3,9 × 108 genome-equivalents per reaction mixture according to the Lukyanov-Matz equation N=2(40-n) , where N – DNA copies before PCR, n – threshold cycle. Taking into account the volume of blood samples 100 µl, incomplete RNA isolation and RT efficiency, one could estimate the DENV loads 107 -1010 virions in 1 ml of a patient blood.
All newborn (1-2 day old) mice in 10-13 days after intracerebral and subcutaneous infection with 10% suspensions of 23 blood grume and lymphocytes of 18 patients (10 of them with DENV-specific antibodies) showed languor, tremor, paresis and growth retardation. The incubation period continued for 7-10 days for the first passage and 6-7 days for the second passage DENV RNA was detected in brain suspensions of the infected mice by RT-PCR. However, part of DENV strains did not appear to be stable with decreased numbers of clinical manifestations on the second passage and the complete absence of observed disturbances during the next third passage but with DENV RNA detection in brain tissue and combined liver + spleen samples in both cases. Therefore, a part of DENV isolates from 48 blood grume and lymphocytes samples from 29 patients (17 of them with DENV-specific antibodies) were recovered on the permissive tissue culture Vero-E6. After infection with the blood samples during 6-8 days Vero-E6 cellular monolayer appeared to gradually destroy from circumference to the center of each well. During subsequent passages the incubation period was shortened up to 4-5 days, in 3-5 passages DENV titers were 5 lg TCID50. In total, among 5 DENV strains (Table 3) one was isolated using mice; three - Vero E6 cells and one – using both mice and Vero E6. To isolate DENV strains, the patient blood samples were taken in 3-4 days of clinical manifestations, in 3 of them the flavivirus-specific or DENV RNA was detected and only one of them was positive in both RT-PCR and ELISA to detect IgM antibodies (Table 2). One should note that 2 DENV strains 2759 and 2996 were isolated from both ELISA and PCR negative blood samples because of possible inhibitors of enzymes in clinical samples, limited sensitivity of RT, or mismatches in primers for PCR that do not correspond to real genetic diversity of new DENV isolates. The discrepancy between three methods of the DENV detection could be explained by their different sensitivity limits and specificity.
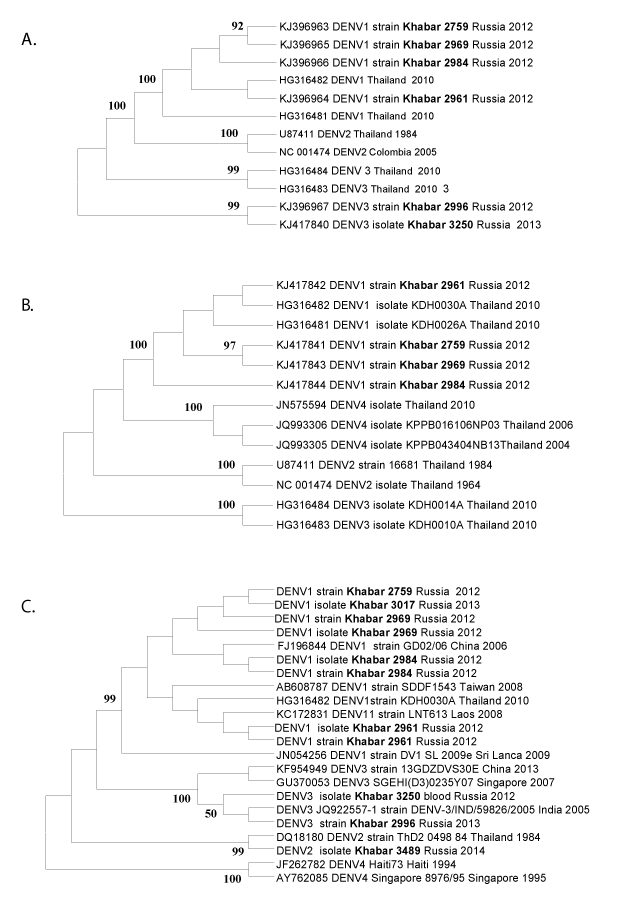
Phylogenetic analysis of nucleotide sequences of 3 fragments of C-prM, E and NS5 genes using Mega 6.06 software revealed similar topologies of phylogenetic trees constructed by using 5 alternative algorithms (Maximum likelihood, Neighbor-Joining, Minimum-Evolution, UPGMA and Maximum Parsimony) and reasonable bootstrap support (part of available data is shown on Figure 1). DENV imported cases were caused by DENV types 1-3. None of the analyzed samples from the patient blood collected in the Far East of Russia appeared to belong to DENV-4 despite well-known co-circulation of DENV multiple serotypes in the South East Asia [5-8]. In Thailand DENV predominant serotype varied year-to-year: DENV-1 from 1990–92, DENV-2 from 1973–86, 1988–89; DENV-3 in 1987 and 1995–99; and DENV-4 during 1993–94 [5,7].
Based on the phylogenetic analysis of DENV E gene nucleotide sequences (Figure 1B) two DENV-1 strains (2759 and 2969) appeared to form a separate clade with excellent bootstrep indexes 97. Nucleotide sequences of C-prM (Figure 1A), E (part B) and NS5 gene (part C) of both strains are also closely related to each other and differ from other DENV strains. One of the DENV-1 strains (2961) isolated in our research was similar to previously described strains from Thailand (Figures 1A and 1C), whereas the strain 2984 was on a separate branch of the trees (Figures 1B and 1C). Direct sequencing of DENV RT-PCR products from patient blood samples completely coincided with the corresponding nucleotide sequences of the virus strains from the infected mouse brain suspensions or from the infected Vero-E6 cells (data not shown). Genetic heterogeneity of any fragment of DENV genomes including the most conserved NS5 gene region [3] (Figure 2A) and the encoding protein diversity (Figure 2B) allowed us to estimate both evolutionary origin and phylogenetic relationships between the DENV isolates (Figure 1).
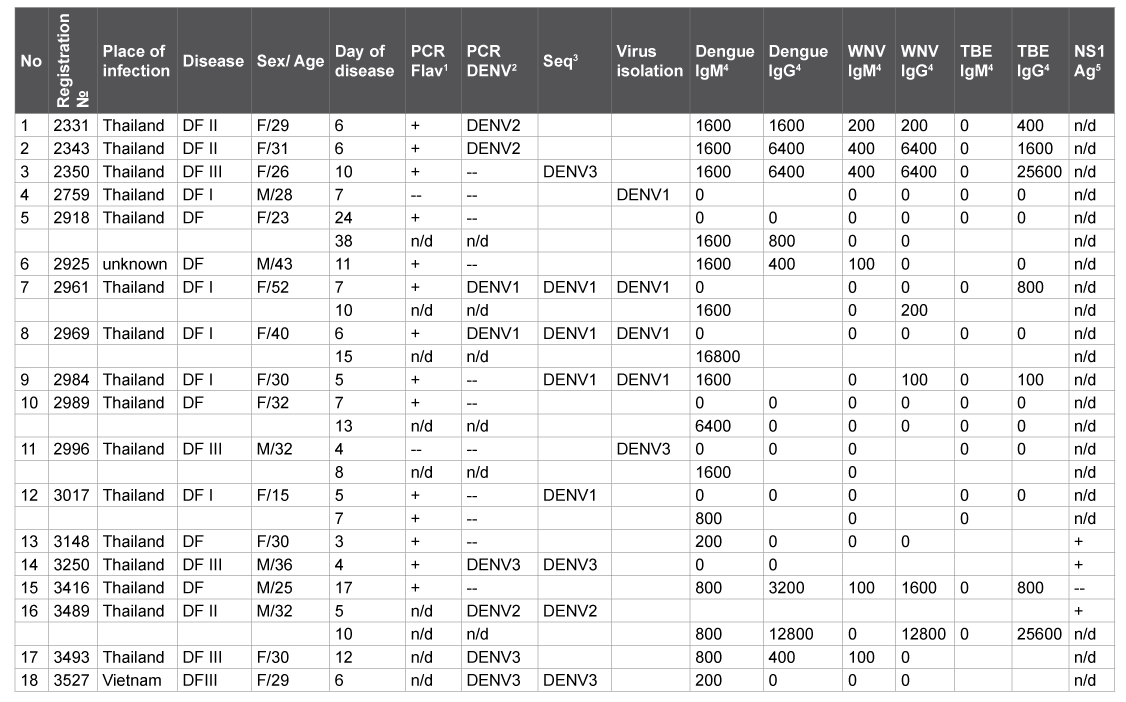
Notes: 1 PCR Flav stands for PCR with universal flavivirus-specific primers (« +» - positive result, «--» - negative result, «n/d» – not done).
2 PCR DENV means PCR with primer pairs specific for DENV 1-4 types.
3Seq – sequencing of the RT-PCR products with universal flavivirus and/or DENV-specific primers.
4 reverse titers in ELISA.
5express test for DENV NS1 antigen.
Table 2: Characteristics of Dengue fever clinical cases (2012-14)
Studies on the imported Dengue Fever cases in the Far East of Russia have corroborated DENV geographic mapping. The majority of DENV strains were isolated from blood samples without specific antibodies [4,9- 11] from patients after their travels to Thailand (Table 3) with the only exception – the DENV-1 strain 2984 from the patient blood with high anti-DENV IgM titer (1:1,600) (Table 2) perhaps due to their low affinity. Besides known limited sensitivity ELISA does not permit to identify viral isolates due to possible immunological cross-reactions. Molecular typing of DENV RNA isolates and strains revealed DENV of 1, 2 and 3 types despite continuous co-circulation of 4 DENV serotypes in Thailand, with one of them resulting in outbreaks [5,7,8].
Besides DENV amplification in human blood with high viral loads in a range 107 -1010 virions per 1 ml the mosquitoes of the genus Aedes can pose an additional threat for the virus transmission in a new endemic region. In the Khabarovsk region 37 species of the mosquitoes of the Culicidae family have been registered: Aedes vexans vexans as a dominant species in southern part of territory with widely spread A. vexans nipponii, A. galloisi, A. cinereus, A. implicates, A. nigrinus, A. flavopictus, A. communius, A. cyptius and other species [16]. A number of viruses have been isolated from the mosquitoes in the Khabarovsk region including TBEV (GenBank accession number KJ744034), Powassan, Negishi, Geta, Batai and the California encephalitis virus [16]. Despite an evaluated likelihood of diagnosing Dengue in a non-endemic region of Russia (nearly 13.7%), an autochthonous spread of DENV is not likely to occur unless global warming makes the region a natural habitat for Anopheles aegypti. Further monitoring of DENV imported cases and molecular epidemiological studies are required.
We thank Doctor Victor F. Larichev (Ivanovsky Institute of Virology, Moscow, Russia) for providing us with ELISA kits to detect DENV-specific IgG and IgM antibodies and Professor Alexander M. Butenko (Ivanovsky Institute of Virology, Moscow, Russia) for his encouragement and support.
DENV strain |
Country of |
Year |
Clinical |
Dengue |
GeneBank accession number |
|
| C-preM | E protein | |||||
| Khabar 2759 | Thailand | 2012 | DF | I | KJ396963 | KJ417841 |
| Khabar 2961 | Thailand | 2012 | DF | I | KJ396964 | KJ417842 |
| Khabar 2969 | Thailand | 2012 | DF | I | KJ396965 | KJ417843 |
| Khabar 2984 | Thailand | 2012 | DF | I | KJ396966 | KJ417844 |
| Khabar 2996 | Thailand | 2013 | DF | III | KJ396967 | n/d |
Table 3: Characteristics of DENV strains isolated in our study

Figure 2: Multiple alignment of DENV NS5 gene fragment nucleotide sequences (part A) and deduced amino acid sequences (part B).
Download Provisional PDF Here
Article Type: Research Article
Citation: Pukhovskaya NM, Morozova OV, Bakhmetyeva SV, Zdanovskaya NI, Belozerova NB, et al. (2015) Molecular Typing of Imported Dengue Virus (DENV) Isolates from Patients in the Far East of Russia. J Emerg Dis Virol 1 (1): doi: http://dx.doi. org/10.16966/2473-1846.101
Copyright: © 2015 Pukhovskaya NM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Publication history:
All Sci Forschen Journals are Open Access