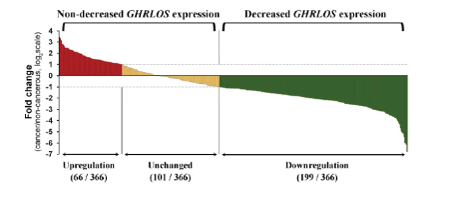
Figure 1: Changes of GHRLSO expression in cancer tissues in 366 patients with colorectal cancer. ***P<0.001.
Xiang Mao Jun Liu# Shuangjie Wu*
Department of General Surgery, Huashan Hospital, Shanghai, China*Corresponding author: Shuangjie Wu, Department of General Surgery, Huashan Hospital, Shanghai, China, E-mail: xmyz1225@aliyun.com
Background: LncRNA GHRLOS is a long non-coding RNAs (lncRNAs). Microarray detection for expression profile revealed that lncRNA GHRLOS was significantly under-expressed in colon cancer tissues, while its role in a tumor remained unclear.
Objective: To explore the function and biological significance of lncRNA GHRLOS in colorectal cancer and elucidate the molecular mechanism of its influence on the malignant biological behavior in colorectal cancer.
Methods: The expression of GHRLOS was detected by collecting the paired tumor tissues and para-cancerous tissues from patients with colorectal cancer. Follow-up data such as recurrence, metastasis, and survival status of patients were collected, and the relationships between the expression level of GHRLOS and indicators such as survival time were analyzed. The expression of lncRNA GHRLOS was changed in colon cancer cells and its influence on migration and invasion of cells was detected. The tumor-bearing model of colorectal cancer in situ in nude mice was constructed to observe the influence of lncRNA GHRLOS expression on metastasis. After the expression of lncRNA GHRLOS was changed, the expression changes of mRNA of the GHRL gene and E-cadherin were detected.
Results: The expression of GHRLOS in colorectal tumor tissues was significantly lower than that in para-cancerous tissues. The down-regulation of GHRLOS expression was associated with lymph node metastasis and distant metastasis of colorectal cancer and resulted as an independent risk factor for the prognosis of colorectal cancer patients. In human colorectal cancer cells, the low expression of GHRLOS causes the cells to become more spindle-like, which negatively regulates the expression of the GHRL gene at the post-transcription level, thus significantly inhibiting the expression of E-cadherin and promoting cell migration and invasion. The overexpression of GHRLOS can inhibit tumor metastasis.
Conclusion: lncRNA GHRLOS can enhance the expression of E-cadherin by directly affecting the mature mRNA of the GHRL gene in colon cancer tissues, which in turn promotes the metastasis, migration, and invasion of tumor cells.
Colon cancer; lncRNA; GHRLOS; E-cadherin
Colorectal cancer is one of the most common malignancies in the world. The incidence of colorectal cancer, which ranks in the top 3 among malignancies in both males and females, is one of the leading causes of tumor-related deaths globally and has been increasing year by year worldwide [1,2]. With the improvement of prevention strategies and early diagnostic techniques aiding in early diagnosis and the optimization of treatment regimens, the survival time and quality of life in patients with colorectal cancer have improved. Yet, there is still a lack of effective treatment measures for metastatic colorectal cancer, resulting in an unsatisfactory prognosis [3]. The metastasis of colorectal cancer is a complex process, which is affected by many biological factors. Currently, there is no unified and comprehensive understanding of its underlying mechanism. Therefore, it is of great academic value to identify the key molecules affecting the metastatic behavior of colorectal cancer and prognosis, which could also have clinical significance for treatments and prognosis.
Long non-coding RNAs (lncRNAs) are a kind of functional noncoding RNA molecules whose transcript length exceeds 200 but lacks a complete open reading frame [4]. For a long time, lncRNA has been considered as a by-product of the transcription process without biological function [5]. In the field of tumor research, the dysregulation of lncRNA expression has been found in a variety of tumors, including colorectal cancer, and has been proved to be closely related to the formation, growth, invasion, and metastasis of tumors [6].
A previous study performed microarray detection of mRNA/ lncRNA expression profiles on the collected tumor tissues and paired para-cancerous tissues of colorectal cancer patients and screened 174 lncRNAs significant changes in the expression of cancer tissues. Meanwhile, 254 differentially expressed mRNAs were obtained. We analyzed the co-expression network of differentially expressed lncRNAs and mRNAs, and further identified seven hub genes in the co-expression network, including lncRNA GHRLOS [7]. In this study, we changed the expression of GHRLOS in colorectal cancer in order to thoroughly investigate its function and mechanism in the process of metastatic behavior of this cancer.
Patients who underwent radical resection of colorectal cancer in the general surgery department of our hospital from 2007 to 2012 were selected. Inclusion criteria were as follows: 1) pathologically confirmed colorectal cancer; 2) firstly received surgical treatment; 3) no preoperative chemoradiotherapy; 4) complete medical records. Exclusion criteria were: 1) unrespectable distant metastatic disease with only palliative surgery; 2) pathological specimens were insufficient to extract RNA; 3) refusal to participate in the study.
A total of 366 patients were enrolled in this study. Tumor tissues and para-cancerous tissues (>5cm from the tumor margin) were collected. All clinicopathological data were collected from medical records. The pathological staging of the patients was based on the AJCC criteria for colorectal cancer staging. This study was approved by the medical ethics committee of Huashan hospital and was implemented in accordance with the Declaration of Helsinki.
Following cell lines that were purchased from the cell bank of the Chinese Academy of Sciences were used in the experiment: SW116, SW480, LOVO, COLO, and 293T. Main reagents included overexpressed lentivirus vectors of GHRLOS-siRNA and GHRLOS, Sigma-Aldrich (Shanghai) Trading Co., Ltd., China; luciferase reporter gene detection kit, RG005, Biyuntian Biotechnology Co., Ltd., China; cDNA synthesis kit, 11123ES60, Shanghai Yisheng Biotechnology Co., Ltd., China; qPCR kit, 1120ES08, Shanghai Yisheng Biotechnology Co., Ltd Technology Co., Ltd., China; BCA protein quantitative kit, c503021, Shanggong bioengineering (Shanghai) Co., Ltd., China; ECL chemiluminescent kit, abs920, Ibixin Biotechnology Co., Ltd., China; HPR-DAB color rendering kit, PA110, Tiangen Biotechnology Co., Ltd., China; HRP labeled goat anti-rabbit, A0208, Biyuntian Biotechnology Co., Ltd, China; HRP labeled goat anti-mouse, A0216, Biyuntian Biotechnology Co., Ltd., China; E-cadherin antibody, 610182, BD Biosciences, USA and N-cadherin antibody, 22018-1-AP, Proteintech, UAS.
After surgery, fresh tumor tissue and para-cancerous tissues more than 5cm away from the margin of the tumor were immediately collected and stored in liquid nitrogen at 80°C for RNA extraction. RNA was extracted with Trizol Reagent (Thermo Fisher technology, China), and the reverse transcription was performed by Primescript RT Reagent Kit (Baori Medical Biotechnology (Beijing) Co., Ltd.). The obtained synthesized cDNA was adopted as a template for quantitative PCR analysis for the expression of lncRNA using FastStart Universal SYBR Green Master Mix (Roche diagnosis, Shanghai). GAPDH was used as a loading control, the Internal Control. The used primers were the following: GHRLOS upstream primer of 5´- TGGAAACTCCCCTAGCCACA; GHRLOS downstream primer of 5´- GCATCTCTCCTCTGTTCCGT; GAPDH upstream primer of 5´- ATCC TGGGCTACACTGAGCACC and GAPDH downstream primer of 5´- AAGTGG TCGTTGAGGGCAATGC.
All enrolled patients were followed up once every three months, and the endpoint of follow-up was October 2016 or death. Followups included physical examination, laboratory examination, imaging examination (endoscopy, ultrasound, CT, MRI, PET) and necessary biopsy, if needed.
The expression of GHRLOS in tumor tissues and para-cancerous tissues was compared by paired t-test. The relationship between GHRLOS expression and clinicopathological features was evaluated by the Chi-square test. The survival curve was drawn by the Kaplan-Meier method, and the differences in the survival curve were analyzed by the time-series test. The Cox regression model was used for univariate and multivariate survival analysis. Variables with p<0.05 in univariate analysis were included in subsequent multivariate analysis. SPSS12.0 was used for data analysis, and the statistical significance level was set at 5%.
Colon cancer cells were cultured in DMEM medium with high glucose in a humidified atmosphere containing 5%CO2/95% air at 37°C. Briefly, cells were cultured with overexpressed GHRLOSsiRNA and lentivirus vectors of GHRLOS and transfection reagent of Lipofectamine®3000 (Invitrogen) at 37ºC, 5%CO2 for 48hours, respectively. The cells were then collected and the morphology of each group was observed. The following cell lines were established: SW116-GHRLOS-NC, SW116-GHRLOS-OE, SW480-GHRLOS-NC, SW480-GHRLOS-siRNA, LOVO-GHRLOS-NC, LOVO-GHRLOSsiRNA, COLO320-GHRLOS-NC and COLO320-GHRLOS-OE (NC: blank control; OE: Overexpression of GHRLOS; siRNA: knockdown of GHRLOS).
Firstly, the virus was transfected into 293T cells, which were cultured in a cell incubator for 72 hours to collect the virus fluid. After concentration, the virus fluid was transfected into colon cancer cells. After the cells were lysed, fluorescein enzyme detection reagent was added to record the absorbance value at 560nm of exciting light.
A total of 5 × 104 cells were seeded in the upper compartment. A serum-free medium was added to the upper compartment, while serum medium was added to the lower compartment. After continuous culturing for 48hours, cell penetration was observed. After formaldehyde fixation and crystal violet staining, the crystal violet was washed out, the upper cells were wiped out, and the number of cells in the lower compartment was observed under the microscope.
Firstly, horizontal lines were evenly drawn on the back of the 6-well plate by a marker, with one line about every 0.5-1cm across the hole and each hole through at least 3 lines. Then, 5 × 105 cells were added to the plate. Twenty-four hours later, the pipette tip was used to scratch at an angle of 90° to the horizontal line on the back. The scratched out cells were removed, and the complete medium was added. A 5%CO2 incubator at 37°C was used for culturing, and photos of the samples were taken at 0, 6, 24, and 48hours.
Cells were digested and centrifuged to collect cells and lysis fluid was added for lysis. The supernatant was collected by centrifugation, which was the total protein solution of cells. BCA kit was used to measure the extracted protein concentration and the protein concentration of all samples was adjusted to 2mg/ml. The SDS-PAGE gel was then prepared. After protein sample was added, electrophoresis and membrane transfer were carried out, after which the primary antibody and secondary antibody were added. Finally, the results were detected by the ECL chemiluminescence detection system.
RNA was extracted with Trizol, while cDNA was synthesized with Hieff® qPCR SYBR Green Master Mix system, and the operation was in accordance with the supplier’s instructions. Primer sequence (5´-3´) was as follows: E-cadherin: upstream primer was TGAAGGTGACAGAGCCTCTGGAT, and downstream primer was TGCGTTCTCTATCCAGAGGCT. GHRL (materal mRNA): upstream primer was AAGATGGAGGTCAAGCAGAAGG, and downstream primer was TCCCAGAGGATGTCCTGAAGAA; GHRL (pre-RNA): upstream primer was GGGCAGAGGATGAACTGGAA and downstream primer was CCTGGCTGTGCTGCTGGTA; GADPH: upstream primer was AGGTGA AGGTCGGAGTCA, and downstream primer was GGTCATTGATGGCAACAA.
The experimental animals were 3-month-old male Balb/c mice of grade SPF (20-25g) that were purchased from Shanghai sipur-bikai experimental animal Co., Ltd., China. All the animals were housed in an environment with a temperature of 22 ± 1°C, relative humidity of 50 ± 1%, and a light/dark cycle of 12/12hr. All animal studies (including the mice euthanasia procedure) were done in compliance with the regulations and guidelines of FuDan University institutional animal care and conducted according to the AAALAC and the IACUC guidelines.
The modeling procedure was as follows: after anesthetizing the nude mice, the abdominal cavity was opened, and the rectum was exposed. A 100μL cell suspension (the total number of cells was 5 × 106) was absorbed with an insulin syringe and inoculated into the colonic serous layer. Small animal imaging was performed every two weeks after tumor cell inoculation. The imaging method was performed as follows: after anesthesia, the nude mice were intraperitoneally injected with 100μL luciferase substrate (at a concentration of 15mg/ml). After waiting for 15 minutes, the nude mice were placed in sequential supine and prone positions in a live animal imager for photographing. After six weeks of feeding, chloral hydrate anesthesia was used to open the abdomen for photographing, and samples were taken for tumor metastasis observation.
The primary and metastatic tissues of the colon cancer model of the nude mice were taken and cut into sections after being trimmed, fixed, dehydrated, and embedded. HR staining and IHC were performed respectively, and E-cadherin was used as the primary antibody.
RT-PCR was used to detect the expression of lncRNA GHRLOS in colon cancer tissues and the paired para-cancerous tissues from 366 surgical specimens. As shown in figure 1, in general, the expression of lncRNA GHRLOS in colon cancer tissue was significantly lower than that in para-cancerous tissues (P<0.001). Among the 366 patients (Figure 2), the expression of lncRNA GHRLOS in tumor tissues in 199 cases (54.4%) was lower than that in paired para-cancerous tissues (multiple of difference
Figure 1: Changes of GHRLSO expression in cancer tissues in 366 patients with colorectal cancer. ***P<0.001. Figure 2: The effect of GHRLOS expression on the prognosis of patients with colorectal cancer by Kaplan-Meier survival analysis.
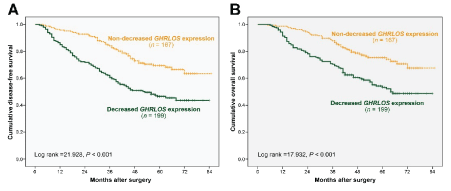
(A) DFS of patients with colorectal cancer in groups with different GHRLOS expression changes by Kaplan-Meier survival analysis.
(B) OS of patients with colorectal cancer in groups with different GHRLOS expression changes by Kaplan-Meier survival analysis.
To determine the role of GHRLOS in the prognosis and survival of patients with colon cancer, the relationships between GHRLOS expression and DFS and OS were evaluated. Of the 366 patients with colorectal cancer, 132 died during follow-ups (Figure 2). Follow-up time after surgery for colorectal cancer patients ranged from 5 months to 85 months, with a median follow-up time of 56 months. The 5-year total DFS and OS for all patients with colon cancer were 57% and 63%, respectively. Survival analysis showed that patients with significantly decreased GHRLOS expression had lower DFS (p<0.001) and OS (p<0.001) than patients with normal GHRLOS expression, thus suggesting that a significant decrease in GHRLOS expression may be a sign of poor prognosis.
To dynamically detect tumor growth in vivo through small animal imaging, we made SW116-GHRLOS-NC, SW116- GHRLOSOE, SW480-GHRLOS-NC, SW480-GHRLOS-siRNA, LOVOGHRLOS-NC, LOVO-GHRLOS-siRNA, COLO320-GHRLOS-NC, and COLO320-GHRLOS-OE cell lines to over-express luciferase. Luciferase activity was detected by luciferase (Figure 3) and the results showed that the above 8 cell lines with luciferase overexpression were successfully constructed. The expression levels of precursor mRNA and mature mRNA of GHRL were detected after knockdown of SW480 and LoVo GHRLOS genes.
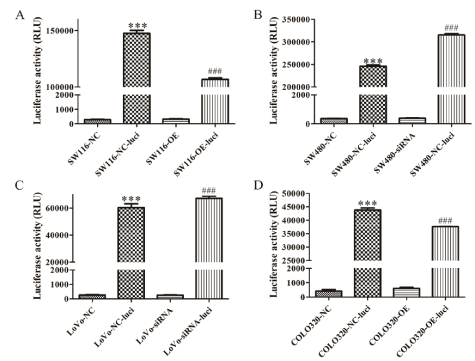
Figure 3: Luciferase was overexpressed in all four cell lines. (A-D) The cartograms of luciferase activity of SW116, SW480, LoVo, and COLO320 before and after luciferase overexpression, respectively. SW116-NC is SW116-GHRLOS-NC, and SW116-OE is SW116-GHRLOS-OE. Similarly, other cell lines omit GHRLOS. Luci represents Luciferase; RLU Represents Relative Light Unit; *** vs NC, P<0.001; *** vs OE/siRNA, P<0.001.
To preliminarily explore the mechanism of GHRLOS regulating GHRL gene expression, the expression levels of precursor mRNA and mature mRNA of GHRL were detected after knockdown of SW480 and LoVo GHRLOS genes. The results showed that after the knockdown of GHRLOS, there were no significant changes in the precursor mRNA expression level of GHRL. In contrast, the expression level of mature mRNA significantly increased (Figure 4), which suggested that GHRLOS may negatively regulate GHRL gene expression at the posttranscriptional level in colorectal cancer.
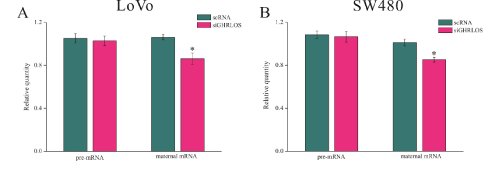
Figure 4: GHRLOS inhibits the expression of mature mRNA of GHRL.
(A) The expression levels of precursor mRNA and mature mRNA of GHRL in LoVo cells before and after knockdown of the GHRLOS gene detected by qPCR.
(B) The expression levels of precursor mRNA and mature mRNA of GHRL in SW480 cells before and after knockdown of the GHRLOS gene detected by qPCR; *, P<0.05.
We observed that after knockdown of the GHRLOS gene, the morphology of SW480 and LoVo cells changed, i.e., they became more spindle-like (Figure 5A and B). Subsequently, the expression levels of E-cadherin in SW480 and LoVo cells were detected, and the results showed that after knockdown of GHRLOS, the expression levels of E-cadherin protein (Figure 5C and D) and mRNA (Figure 5E) both significantly decreased, which suggested that GHRLOS may have an anticancer role by inhibiting EMT in colorectal cancer.
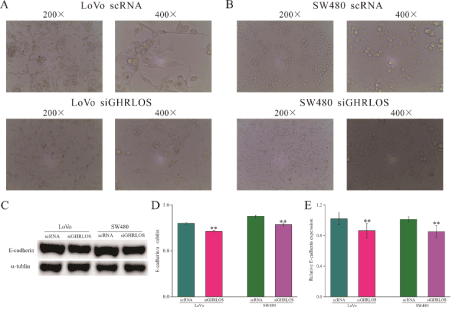
Figure 5: Low expression of GHRLOS promotes EMT in human colon cancer cells.
(A-B) The morphological changes of LoVo and SW480 cells before and after knockdown of the GHRLOS gene observed by microscope, respectively.
(C-D) The effect of low expression of GHRLOS on the expression level of E-cadherin protein in LoVo and SW480 cells detected by WB.
E shows the effect of low expression of GHRLOS on the expression level of E-cadherin mRNA in LoVo and SW480 cells detected by qPCR; *, P<0.05; **, P<0.01.
Scratch test and transwell experiment revealed that the migration and invasion ability of SW480 and LoVo cells significantly increased after knockdown of the GHRLOS gene (Figure 6).
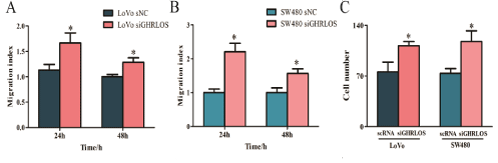
Figure 6: Low expression of GHRLOS promotes migration and invasion of human colon cancer cells.
(A-B) The cartograms of migration ability of LoVo and SW480 before and after knockdown of the GHRLOS gene.
(C) The cartograms of invasion ability of LoVo and SW480 before and after knockdown of GHRLOS gene; *, P<0.05.
The overexpression of GHRLOS promoted the expression of E-cadherin and inhibited the expression of N-cadherin in tissues of the primary lesions, thus impeding the formation of metastasis.
To further verify whether GHRLOS also has an anti-tumor role in vivo, we constructed a tumor-bearing model of colorectal cancer in situ in nude mice using SW116 and SW480 cells. Small animal imaging was performed every two weeks. IHC results of the primary (colon) lesion also showed that the overexpression of GHRLOS promoted the expression of E-cadherin and inhibited the expression of N-cadherin (Figure 7). In the 6th week, the imaging results of small animals and the final anatomical results of nude mice showed that GHRLOS overexpression could inhibit the formation of cancer metastasis (Figure 8). All the suspected tumor metastases were verified by HE staining, and the results showed that all of them were cancerous (Figure 8). These results indicated that GHRLOS overexpression could inhibit EMT and tumor migration.
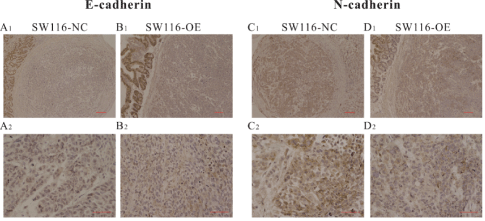
Figure 7: GHRLOS overexpression promoted the expression of E-cadherin and inhibited the expression of N-cadherin in the primary lesion of colon cancer.
(A-B) The expression of E-cadherin in the primary lesion of colon cancer detected by IHC.
(C-D) The expression of N-cadherin in the primary lesion of colon cancer detected by IHC; A1-D1 scale represents 100 μm; A2-D2 are the local enlarged images of A1-D1, and the scale represents 50 μM.
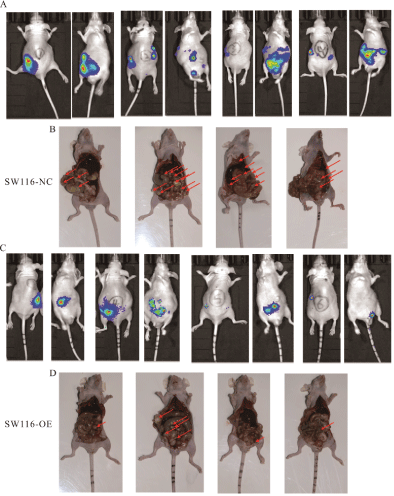
Figure 8: GHRLOS overexpression inhibits metastasis of colon cancer.
(A) Small animal imaging results of the SW116-NC group at 6th week.
(B) Anatomical drawing of the SW116-NC group at the 6th week of execution and sampling.
(C) Small animal imaging results of the SW116-OE group at 6th week.
(D) Anatomical drawing of the SW116-OE group at the 6th week of execution and sampling; The red arrow indicates the location of the tumor.
LncRNAs are believed to have an essential role in the biological behavior of tumors. The expression variation of lncRNA is involved in the development of tumors and may be a potential prognostic marker for patients [8]. In colon cancer, the abnormal expression of many lncRNAs, such as HOTAIR [9], NEAT1 [10], and H19 [11], are believed to be related to the prognosis of colon cancer. In our previous study, the expression profiles of colon cancer tissues and paired paracancerous tissues were compared, revealing many differentially expressed lncRNAs and mRNAs [12]. Further co-expression analysis of these abnormally expressed lncRNAs and mRNAs revealed that lncRNA GHRLOS was one of the hub genes of the lncRNA/mRNA co-expression network in colon cancer [12]. However, the clinical significance of GHRLOS in colon cancer is still poorly understood.
In the present study, we analyzed GHRLOS expression in 366 pairs of colon cancer tissues and paracancerous tissues. Our results revealed that in tumor tissues, GHRLOS expression was decreased in a large proportion, thus suggesting that GHRLOS had an important role in the evolution of colon cancer. Furthermore, it was found that the downregulation of GHRLOS expression was positively correlated with poor prognosis in colorectal cancer patients. The above results suggested that GHRLOS may have the function of tumor suppressor genes and that down-regulation of GHRLOS may promote the progression and metastasis of colorectal cancer. Therefore, detection of GHRLOS expression can help identify patients with a high risk of recurrence of colorectal cancer. Consequently, it is necessary to evaluate whether this population needs more aggressive treatment.
Clinically, the down-regulation of GHLOS expression was positively correlated with the poor prognosis of colorectal cancer patients. In view of cell biological behavior, our results revealed that the low expression of GHRLOS caused the cells to become more spindle-like. Furthermore, they could promote cell migration and invasion, while the overexpression of GHRLOS promoted the expression of E-cadherin and inhibited the expression of N-cadherin in tissues of primary lesions, which impeded the formation of the metastases. However, after knockdown of GHRLOS, the expression level of mature mRNA of GHRL significantly decreased, thus indicating that GHRLOS may regulate the expression of the GHRL gene at the post-transcriptional level in colorectal cancer. In conclusion, GHRLOS may affect the EMT process of tumor cells by regulating the expression of the GHRL gene, thus affecting the migration, invasion, and metastasis of cells.
Since GHRLOS is an antisense RNA of the Growth HormoneReleasing Peptide (GHRP) gene, many scholars believe that GHRLOS has a vital role in regulating the auxin axis [13]. GHRP is an important gastrointestinal peptide hormone, which is the ligand of growth hormone-releasing receptors [14]. It has an essential role in the regulation of energy metabolism but is also deeply involved in the growth and progression of many tumors [7,14,15]. In colorectal cancer, the overexpression of GHRP was observed in both tumor cell lines and pathological tumor tissues [16]. The increase of GHRP was also found in the serum of patients with colon cancer, supporting the hypothesis that tumors secrete GHRP. Previous studies have confirmed that GHRP could promote the proliferation, migration, and invasion of colorectal tumors [16,17]. The downregulation of the GHRP receptor could inhibit the growth of colorectal cancer cells in vitro and in vivo. Since GHRLOS is the antisense RNA of the GHRP gene, it is possible that the down-regulation of GHRLOS could lead to the activation of the signaling pathway of GHRP and promote the growth and metastasis of colorectal tumors. However, the regulatory mechanisms of GHRLOS expression are not fully elucidated, and the transcription factors regulating GHRLOS expression have not yet been found.
In general, our results revealed that most of the expression of lncRNA GHRLOS was downregulated in colon tumor tissues. Clinically, IncRNA GHRLOS is not only expected to serve as a biomarker to predict metastasis, but also to predict the prognosis of patients. At the cell level, the low expression of GHRLOS causes cells to become more spindle-like, and to promote the migration and invasion of cells. In contrast, the overexpression of GHRLOS promotes the expression of E-cadherin and inhibits metastasis in tissues of primary lesions. At the molecular level, after knockdown of GHRLOS, the expression level of mature mRNA of GHRL was significantly decreased, which may be the intrinsic mechanism of GHRLOS to exert biological functions. The reported observations need to be further confirmed by future experimental studies.
LncRNA GHRLOS can enhance the expression of E-cadherin by directly acting on the mature mRNA of the GHRL gene in colon cancer tissues, which in turn inhibits the metastasis, migration, and invasion of tumor cells.
None declared.
Download Provisional PDF Here
Article Type: RESEARCH ARTICLE
Citation: Mao X, Liu J, Wu S (2021) Inhibitory Effect of lncRNA GHRLOS on Colon Cancer Metastasis and its Molecular Mechanism. J Surg Open Access 7(4): dx.doi.org/10.16966/2470-0991.251
Copyright: © 2021 Mao X, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Publication history:
All Sci Forschen Journals are Open Access