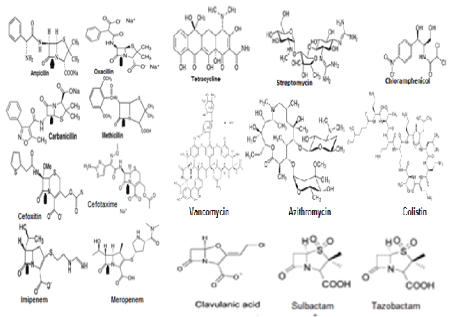
Figure 1: Structures of diverged antibiotics that are now useless in superbug infections [14].


Asit Kumar Chakraborty*
Department of Biochemistry and Biotechnology, Oriental Institute of Science and Technology, Vidyasagar University, Midnapore, West Bengal, India*Corresponding author: Asit Kumar Chakraborty, Department of Biochemistry and Biotechnology, Oriental Institute of Science and Technology, Vidyasagar University, Midnapore-721102, West Bengal, India, Tel: 91+9339609268; E-mail: chakraakc@gmail.com
AMR spread is huge with millions of death due to ineffectiveness of antibiotics. This is happened due to creation of hundreds of mdr genes like beta-lactamases (blaTEM, blaCTX-M, blaOXA and blaNDM1), drug acetyltransferases (aacC1, aacA1, cat) and phosphotransferases (aph4) in MDR conjugative plasmids and chromosome. In other mechanisms, drug efflux proteins like tetA/C, acrAB and mexAB kickout drugs from bacterial cytoplasm increasing drug MIC and thus tetracycline, streptomycin, azithromycin, ciprofloxacin became ineffective to destroy pathogenic bacteria. We have investigated here the reason of such widespread creation of mdr genes in bacteria which are resident of intestine synthesizing 20 vitamins and complex biomolecules for our body. We have hypothesized that such 2x1012 diverse species are killed due to high dose of antibiotic intake since 1940s creating acute health hazard in human which is now balanced by probiotic bifidobacteria and vitamin-B complex capsules supplement. The hypothesis suggested that molecular signalling from intestinal luminal cells (TGFβ, IL-10, IL-22) as well as from bacteria (LPS, vitamins, butyrate) orchestrated to preserve symbiosis relation between human and bacteria. Vitamins are converted into provitamins (FADH2, NADH+, THFA, Biotin, B12, TPP etc.) needed for every steps of >30000 enzymatic reactions in human cells. Thus MDR bacteria will be the resident of intestine favouring vitamin biosynthesis and immune-modulation needed for normal human metabolosome. Indeed mdr genes are abundantly created in plasmids and chromosomes with further mutations of target genes (rRNA, ponA, porB, gyrAB, parC) and likely all are to protect gut microbiota to save human from extinct. Thus with time all bacteria will be drug resistant and infections should be controlled by heterogenous phyto-antibiotics, gene medicines, phage therapy and DNA nanocarriers for toxic drug delivery.
AMR; Antibiotic void; Gut microbiota; Vitamin synthesis; Gene rearrangement; MDR genes
The existence of microbes were described until Von Leeuwenhoek observed microorganism through his microscope in 1680s followed by eminent microbiology work of Louis Pasteur and Robert Koch in the 1850s [1]. The antibiotic principle was discovered in 1926 by Alexander Flaming but it took until 1943 for large scale production and supply of penicillin drug for peoples [2]. So mass peoples were taken antibiotics after World War II and within last 60 years we eradicated gut microbiota so drastically that save your soul worked signalling from human and bacteria both to create mdr genes many fold. It is unbelievable to thing that penicillin, streptomycin, tetracycline, chloramphenicol, refampicin, azithromycin, ciprofloxacin, cefotaxime, sulfamethoxazole, trimethoprim, are not killing bacteria (Figure 1 for antibiotic structures). Those bacteria are termed as superbugs as highly resistant to at least three groups of different antibiotics. All antibiotic groups are different structures and their derivatives are sometime gave unique side chains producing better drugs but all in vain as ultimately drug resistant genes modified due to mutations or a new isomers appeared that destroy the antibiotic easily than expected (Figure 2 for many MDR Genes). Dr. Selman Walksman’s streptomycin worked very well to clear TB in 1950s but now XRD-TB increased many fold in India and abroad. MDR Mycobacterium tuberculosisis drug resistant today with strA/B, arr3, katG mdr genes and mutations in the ribosomal genes rpsL, rplC and rrs as well as pncA gene involved in pyrazinamide resistance [2,3].

Figure 1: Structures of diverged antibiotics that are now useless in superbug infections [14].
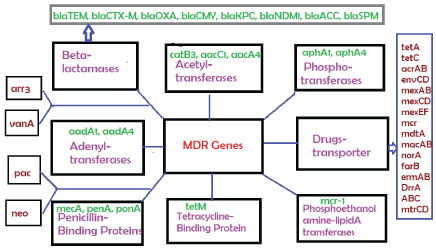
Figure 2: Classification of diverged multi-drug resistant (mdr) genes that destroy all antibiotics described in Figure 1 by different mechanisms.
The length of conjugative plasmids are expected 70-80kb (62kb F’- plasmid + 5-15kb R-plasmid) but now many recombinations have produced 100-500kb plasmids with 5-15 mdr genes, >10-20 Tra conjugative genes and other 10-20 new genes whose functions remains to be elucidated [4]. Surely we find toposiomerase III, parA, Uvr3, parC, Transposons and ISelements (Tn3, IS-26), ABC transporters and metal resistant genes (terA/ B/C, CopA, MerB/C/X) in most MDR conjugative plasmids (Figure 3 for structure of prototype large MDR conjugative plasmid in superbugs). A 150kb IncA/C plasmid pMRV150 in Vibrio cholerae 0139 strain was found resistant to common antibiotics, ampicillin, tetracycline, gentamycin and chloramphenicol. A IncC hybrid 165kb plasmid in Proteus mirabilis was discovered in 2017 with 15 mdr genes including most deadly blaNDM-1 and blaCTX-M-65 and indicated that how severe recombination was facilitated in the human intestine during antibiotics exposure [5-8].
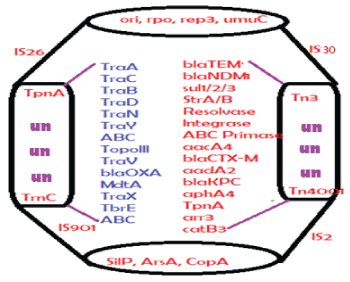
Figure 3: Structure of a large MDR conjugative plasmid. Such plasmids present in superbugs containing 10 mdr genes and 20 conjugative genes including metal resistant genes and many IS-elements and transposons involved in gene transfer and rearrangement. The plasmid accession numbers are: NC_018107b (353kb), NC_022078 (317kb), LN555650 (299kb), KC543497 (501kb), JN420336 (267kb), CP007558 (272kb), CP011634 (227kb)and FJ628167 (151kb). Un means unknown genes and Tra is conjugative genes.
The drug industry is always run to make better derivatives of existing drug like Benzyl penicillin. So ampicillin and amoxicillin semi-synthetic drugs were prepared in 1960s to overcome the action of penicillinase enzyme (amp gene, discovered in Escherichia coli cloned plasmid pBR322 and sequenced in 1965) [2]. However, blaTEM-1 (protein id. AAB59737) and blaSHV (protein id; AAD37412) enzymes inactivate ampicillin. So oxacillin derivatives were prepared but soon blaOXA-1 enzyme (protein id: AFG30109) was developed that destroy oxacillin and ampicillin but Class-D higher beta-lactamase derivatives like OXA-23, OXA-48 were more potent and most beta-lactams were inactivated. Similarly, blaCTX-M (protein id: ABN09669) efficiently hydrolyses all derivatives of cehalosporins including cefoxitin, cefotetan, ceftriaxone and cefotaxime. In 2009 blaNDM-1 beta-lactamase (protein id: AGC54622) was discovered in Escherichia coli and Klebsiella pneumoniae plasmids. Such enzyme hydrolyses the potent carbapenem drugs like imipenem, dorripenem including cefotaxime and ampicillin drugs (see, Figure 4 for gradual discovery of penicillinases, cephalosporinases and carbapenemases) [9].
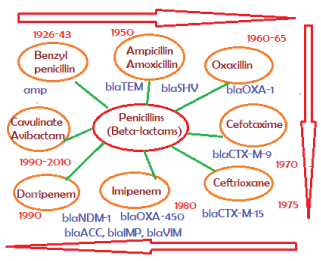
Figure 4: Discovery of β-lactams and β-lactamases. It appears any insult of gut microbiota by antibiotics will follow drug resistant with new mdr gene synthesis in the intestinal bacteria. About 20 types distinct β-lactamases with few thousand mutations were detected and. Iit appears bla genes are is also acquired enhancer-repressor elements and are bla genes are now induced by β-lactams. So use of more antibiotic means more beta-lactamase synthesis increasing MIC and is likely health risk [4].
Most bacteria in sea, river and rain water are not only resistant to betalactams but also to tetracycline, streptomycin, vancomycin, azithromycin and so many others (see Figure 5) [10,11]. However, tetracycline resistant determinants are also many fold: (a) MFS drug efflux type enzymes; tetA (protein id: CAA53389), tetC (protein id: AGL61405) (b) tetracycline binding proteins like tetM, tetO, tetS that induce ribosome protection and (c) tetracycline modification enzyme like tetX that acts in presence of O2 and NADPH [2]. We characterized the Kolkata environmental superbugs that located in drain water, street rain water, Digha sea water (Bay of Bengal) and most importantly Ganga River water and all were resistant to ampicillin and amoxicillin as well as gentamycin, streptomycin and ciprofloxacin [11,12]. As imipenem, vancomycin, amikacin, colistin, ceftriaxone lomofloxacin and azithromycin resistant species were frequent, it help us to thought differently (see, Figure 5 and Table 1). Such conclusions are immerged after carefully studying the multiple mdr gene isomers at the genetic label and drug sensitivities which shows obscure and unbelievable pressure likely generated in vivo to create mdr genes with all possible combinations of protein primary amino acid sequences (see, Table 1) [4,13,14]. We have discussed here the rationally of antibiotic void creating an antibiotic dark age due to overuse of antibiotics. We have pinpointed the odd that we misconceptually have killed all intestinal symbiotic bacteria that help us to life and now we need alternate to antibiotics for better treatment of MDR bacterial infections.
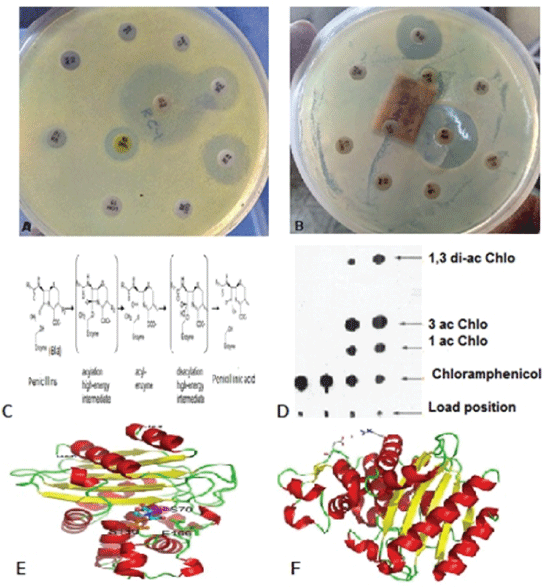
Figure 5: (A-B) Multidrug resistant assay using HI-Media antibiotic papers; (C) cleavage mechanism of beta-lactam by beta-lactamase enzyme; (E) CAT gene assay to demonstrate acetylation of chloramphenicol and (E-F) crystal structures of blaTEM and blaKPC enzymes [4,14].
| Antibiogram of the isolated and characterized MDR-bacteria from Kolkata | ||
| Bacteria/Source | Accession no | Major Drug Resistance Patterns |
| Escherichia coli KC-1 mdr (Ganga river) | KU898253 | Methicillin, Tetracycline, Streptomycin, Ciprofloxacin, Cotromoxazole, Lomofloxacin, Vancomycin, Amikacin, Linezolid |
| Escherichia coli KR-1_mdr (Kolkata rain) | KY769883 | Methicillin, Cefotaxime, Aztreonam, Azithromycin , Chloramphenicol, Tetracycline, Vancomycin, Amikacin, Streptomycin, Cotrimoxazole |
| Pseudomonas aeguginosa DB-1 mdr (Kolkata street) | KY769875 | Metthicillin, Cefotaxime, Ceftrioxane, Azithromycin, Chloramphenicol, Vancomycin |
| Escherichia coli KC-2_mdr (Kolkata street) | KY769878 | Methicillin, Tetracycline, Amikacin, Vancomycin, Cotrimoxazole, Streptomycin, Azithromycin |
| Escherichia coli KT-1_mdr (Kolkata street) | KY769881 | Metthicillin, Cefotaxime, Tetracycline, Ciprofloxacin, Chloramphenicol, Azithromycin, Gentamycin, Vancomycin, Amikacin, Linezolid |
| Escherichia coli KT-2_mdr (Ganga river) | KY769882 | Metthicillin, Vancomycin, Lomofloxacin, Cotrimoxazole, Azithromycin, Amikacin |
| Phenalkaligenes sp. KG-1_ mdr (Ganga river) | KY769879 | Neomycin, Gentamycin, Azithromycin, Ciprofloxacin, Polymyxin, Vancomycin |
| Stenotrophomonas sp. KGB- 1_mdr (Ganga river) | KY769880 | Methicillin, Azithromycin, Tetracycline, Streptomycin, Cotrimoxazole, Vancomycin |
| Escherichia coli KTB-1_mdr (Kolkata street) | KY769877 | Methicillin, Tetracycline, Cotrimoxazole, Neomycin, Streptomycin, Azithromycin |
| Pseudomonas aeruginosa DG-2_mdr (Digha sea) |
KY769876 | Cefotaxime, Ciprofloxacin, Tetracycline, Streptomycin, Cotrimoxazole, Neomycin |
Table 1: Antibiogram and 16S rRNA sequencing to confirm MDR bacterial Genus and species. rRNA genes were deposited into NCBI GenBank [30]. All bacteria are resistant to ampicillin but sensitive to imipenem. Imipenem resistant bacteria was recovered from 10 ml Ganga River water and Digha sea water but was absent in rain water [2].
Water from Ganga river was collected at Babughat (Kolkata 700001), at Howrah station ghat (Howrah 711101) and Rain water was collected at South Kolkata (Kolkata 700032). 100μl water spotted onto LB-1.5% plates in presence of single or all four antibiotics cocktails containing tetracycline, choramphenicol, ampicillin, azithromycin, ciprofloxacin and streptomycin. Individual colony was picked up and was grown in presence of antibiotic (ampicillin). The water from Ganga River, Kolkata streets and famous railway stations after rain in monsoon seasons (June– July) gave very distinct drug resistant colonies in presemce of 50µg/ml of ampicillin and amoxicillin with 4500-5500 cfu/ml of water [12]. The cfu/ml of water were reduced to five fold in case of tetracycline and azithromycin at 20µg/ml and 50µ/ml respectively, where as reduced to 49 fold with 34µg/ml chloramphenocol and 50µg/ml streptomycin. In presence of beta-lactamase inhibitors cavulinic acid and sulbactam, cfu/ ml further reduced to ~50 cfu /ml of water. Similarly inclusion of three antibiotics tetetrcycline, ampicillin, chlormphenicol, streptomycin in combination produces only 10-20 cfu/ml water. Further, imipenem resistant species were found rare with only 0.2-0.3 cfu/ml of Ganga River water.. The results indicated that everywhere had MDR-bacteria and 30-40% was ampicillin and amoxycillin resistant as well as <1-2% were superbugs (MDR but % XDR was low and no PDR was detected). As for example, imipenem resistant bacterial species were present extremely low (~ 0.003%). According to law, MDR bacteria must be resistant to at least three different groups of antibiotics. So the percentage of MDR-bacteria that resistant to three drugs, ampicillin, streptomycin and tetracycline was also as low as only 0.2%. Then we have tested the pure rain water (collected on 4th floor roof keeping a 50 ml plastic tube inside a 500 ml beaker) and it has also very similar numbers of bacteria indicating as the major source of bacterial contamination on streets, ponds and sea. This means if any superbug got escape from clinics to environment by physical calamity like storm, tide, flood or earth quake and then bacterial spore could be spread to anywhere by wind and would fall during rain affecting mass populations. The old city like Kolkata has damage sewage system and floods everywhere of the city during monsoon causing nail or skin infections. We have isolated few strains of gram-negative superbugs (KA1, KR1, DG1, KC1, KT1, and KG1 that are resistant to at least three different groups of drug (e.g. ampicillin, streptomycin, cefotaxime, azithromycin, ciprofloxacin, tetracycline or chloramphenicol). The nomenclature has given as follows: “K” means Kolkata origin, KA1 means ampicillin selected 1st then mdr-selected; KR1 means rain source and mdr-selected, KC1 means 1st chloramphenicol selected then mdr selected; KT1 means 1st tetracycline selected then mdr-selected, KG1 means from Ganga river water. Mdr-selection usually mean LB-agar plate contains a mixture of four antibiotics at 50-100µg/ml concentration (ampicillin, streptomycin, ciprofloxacin, azithromycin. The bacterial counts in water sources from open drain, rain water and Ganga River at Kolkata were compared and surprisingly high incidence (~ 40%) of penicillin drug resistant bacteria were found everywhere. Most of the bacteria were Escherichia rod and flagellates and Pseudomonas as demonstrated by electron microscopy and also could form circular spores. Surprisingly, KG-1, KT-1 or KC-1 strains are resistant to 12 Hi-Media antibiotic strips according to CLSI standard [11].
The genomic DNA was isolated from 3 ml ON culture in LB media (10gm NaCl+10gm Bactotryptone+5gm Yeast extract/L water at PH 7.4) in presence of 100µg/ml ampicillin or 40µg/ml tetracycline. The bacteria were pelleted at 5000 rpm and pellet was dissolved in TE buffer and incubated overnight in presence of 0.1% SDS and 20µg/ml Proteinase-K. Then extracted with chloroform: isoamyl alcohol (24:1) and precipitated with 2 volumes of 99% ethanol. Genomic DNA was dissolved in TE buffer and treated with DNase free RNase A (1µl of 20mg/ml) for 15 min at 37ºC, extracted with phenol: chloroform: isoamyl alcohol (25:24:1) and DNA precipitated with 1/9 volume 3M sodium-acetate PH 5.2 and 2 volume ethanol.
The plasmid DNA was isolated from overnight culture using AlkalineLysis Method. Simply, to bacterial pellet 100 solution-I was added and mixed. Then 200µl of cold Solution-II added to make transparent solution and then 150µl cold of Solution-III was added and mixed well. After 10 min the solution containing huge white coloured precipitate of chromosomal DNA-cell debries were removed by centrifugation at 10000rpm for 10min.. To clear solution then added 1 ml 99% ethanol and centrifuged at 10000 rpm for 10 min att 4ºC. Plasmid DNAs from four such preparation were combined and the tRNAs were removed by Rnase A treatment as above and finally plasmid DNA was dissolved in 50µl TE buffer and was stored at -20ºC. 0.8% agarose gel electrophoresis in 1x TAE buffer at 50V for 4-6 hrs was performed to see the plasmid DNAs after staining in 0.5µg/ml ethidium bromide and UV illumination [11].
| Primers used in to detected beta-lactamase and drug efflux genes | |||
| Name | Sequence of the primers | Tm | size |
| P27F | 5’-AGA GTT TGA TCC GAA CGC T-3’ | 62ºC | 1.4kb |
| P1392R | 5’-TAC GGC TAC CTT GTT ACG ACT TCA-3’ | 65ºC | |
| cmrF | 5’-TTC GTT AGT CTG CCG TTG CT-3’ | 56ºC | 323bp |
| cmrR | 5’-ATC GCT GGC AAA CAG GGT TA-3’ | 57ºC | |
| blaVIM-F | 5’-CAG ATT GCC GAT GGT GTT TGG-3’ | 57ºC | 519bp |
| blaVIM-R | 5’-AGG TGG GCC ATT CAG CCA GA-3’ | 61ºC | |
| tetF | 5’-CTT CGC TAC TTG GAG CCA CT-3’ | 57ºC | 910bp |
| tetR | 5’-GCA GAC AAG GTA TAG GGC GG-3’ | 57ºC | |
| acrAB-F | 5’-ATG CTC TCA GGC AGC TTA GCC-3’ | 59ºC | .1kb |
| acrAB-R | 5’-TGT CAC CAG CCA CTT ATC GCC-3’ | 59ºC | |
The 1.4 kb rRNA DNA amplified by sequence specific forward primer 5’-AGA GTT TGA TCC GAA CGC T-3’ and reverse primer 5’-TAC GGC TAC CTT GTT ACG ACT TCA CCC C-3’ using Taq DNA polymerase. The PCR reaction contained (30µl) 50ng genomic DNA, 0.25mM dXTPs, 2mM MgCl2 and 1Unit Taq polymerase, and amplification cycles were 35 for 94ºC/30sec, 52ºC/45sec, 72ºC/2min. The 1.4 kb band was excised from gel and 20-50 ng DNA was used for di-deoxy colour chain termination reaction for DNA sequencing by Xcelris Labs Limited and SciGenom Labs.
Sequence obtained by forward and reverse primers were aligned and homologies were detected by BLAST (www.ncbi.nlm.nih.gov/blast). Then the bacteria was recognized for genus and sometime species ana deposited to GenBank and accession numbers were obtained (KY769875- KY769883). The papers and reviews were obtained using Pubmed (www. ncbi.nlm.nih.gov/pubmed) and individual DNA/Protein sequence (plasmids and genomes and enzymes) was obtained by nucleotide/protein search (www.ncbi.nlm.nih.gov/nucore//nucleotide//protein). Individual mdr gene was sorted and seq-2 DNA analysis was performed to detect the chromosomal and plasmid position. Then the individual accession number was searched for DNA sequence of plasmid and chromosome to check the ATG and TAA codons [2,14].
We confirmed in 1940s that we got magic bullet “antibiotics” and we could withstand World War I and II madness thereafter with removing bugs from our body by Flaming’s and Walksman’s magic bullets “penicillin and streptomycin”. In 2017, we confirmed that all antibiotics had created problems in our body by signalling to make mdr genes highly so that repeated dose of antibiotics would not able to kill all microbiota in our intestine. It seems bacteria are very happily rearranged its DNA to make 100 of mdr genes in plasmids and integrons which now have recombined with F’-plasmid to make super conjugative MDR plasmids that could donate mdr genes easily to all bacteria. Thus >95% of bacteria isolated from our body are now ampicillin and tetracycline resistant [11,12].
Table 1 shows the antibiotic sensitivities of MDR bacteria isolated from Kolkata water resources and drug resistant is rampant. Scientists have predicted that mdr genes could be present in bacteria before the discovery of antibiotics in 1928 due to competition between bacteria and fungi [15]. We assume such genes are very similar to modern amp, tet, neo, blaNDM-1, blaOXA-23, blaCTX-M-1, aacA4 (protein id: AEZ05102), aacC2 (protein id: AAA21890), aphA2 (protein ids: CAA25854, AAA85506), aadA1 (AAK13440), sul1, arr, mcr-1 (protein id: ARD68168), catB3 (protein id: AAD20921) etc. genes that are generated due to over exposed antibiotics in human and animal as well as contamination of such drugs in water from industry, agricultural land and human excreta (Figure 6) [3-5]. Question arises if bacteria are needed for human development is known, then why drastic antibiotic use is permitted by physicians to remove gut micro-flora? Of course pathogenic bacteria should be eliminated by antibiotics but probiotic bacteria should be taken instantly after each antibiotic exposure. Now >40% sea and river water Escherichia coli and >95 % clinical isolates are resistant to ampicillin and amoxicillin, the wonder drugs that are used since 1943 [16-18]. Why not scientists have tried to understand that 20 vitamins and many complex bio-molecules are produced by bacteria without which we cannot live one minute because coenzymes are needed for glycolysis, TCA cycle, ATP generation and metabolisms of DNA, RNA, protein and lipid [19-21]. Vitamin-B and vitamin–A complex are approved for treatment but few peoples know about it and doctors are also not very keen to give message to patients about it particularly in poor nations. Recently, researchers have suggested that reduction of antibiotics use will lower the spread of drug resistant bacteria and G-20 action plan on AMR tells the truth. Moreover, several high quality research from US Human Microbiome Project (HMP), European Metagenomics of the Human Intestinal Tract (MetaHIT) and others have demonstrated the beneficial functions of the normal gut flora (>35000 species) on health (Figure 6). Such microorganisms express many hydrolases, glycosyl transferases and polysaccharide lyases as in Bacteroides thetaiotaomicron about 260 hydrolases have been reported that even are not present in 23 pair human chromosomes [22]. Similarly other microorganisms like Bacteroides, Roseburia, Bifidobacterium, Fecalibacterum and Enterobacterium are involved in carbohydrate metabolism with production of butyrate, acetate and propionate nutrients that are involved in molecular signalling to intestinal cells (Figure 7) [23]. Other study indicated that oxalate was removed by Oxalobacter formigenes and Lactobacillus species preventing kidney stones. Intestinal Escherichia coli and Bacteroides intestinalis have role in modulation of bile acids into deoxycholic acids and lithocolic acids [22,23]. Similarly, pathogenic bacteria like Helicobacter, Vibrio, Salmonella in stomach, Enterococcus, Bacteroides fragilis in luminal, Clostridium sp., Prevotella sp. and Akkermansia muciniphila in intestinal mucus layer are important in bioconversion, vitamin biosynthesis and immunemodulation (Figures 7 and 8) [13]. Nevertheless, roles of Helicobacter pyroli and Pseudomonas aeruginosa in promoting cancer have been established and in peoples mind first and last thing is take sufficient antibiotics to clear unseen stomach and intestinal bacteria (Figure 6). Technology like microbial culturomics help to detect and isolate previously uncultured gut bacteria for typing against hundred of antibiotics and bacteriophages that again may facilitate MDR generation with new gene creation [14]. Gut microbiota populations greatly vary with age but most bacteria could be found within age 3 and changes in such diverse populations cause serious health hazards like obesity, cancer, diabetes and others (Figure 6). Rapid and repeated destruction of such microbiota by antibiotic uses since 1940s have caused serious commucations gaps by LPS and vitamin synthesis. Now more and more new genes will be seen in plasmids (each 100-500kb MDR conjugative plasmid carry ~20 unknown genes) as rapid DNA sequencing methods by Illumina (SanDiego), Roche 454 pyro-sequencing, HTS technology like solid system (Applied Biosystems), and Ion platforms (Life Technologies) are available to all researcher with a reduced cost [24,25]. No doubt free cost of NCBI database and BLAST search technologies now have facilitated bacterial genome sequencing at every college and research institution of the developing countries (Table 2 for plasmid accession numbers). We also know that without amp, neo cat, aac, pac genes no vector could be devised and similarly few thousand vectors are used everyday where huge antibiotic are used for recombinant bacterial selection. So ampicillin, streptomycin, chloramphenicol, neomycin and tetracycline are used few decades largely increased the concentration of drugs into environment. Critic argue that such bacteria (E. coli DH5 α) have genetic mutations and uncapable of conjugation and recombination [11-13]. My point is scientific development favours industry and business benefiting developed countries but poor countries have to face problems due lack of infrastructure and sufficient fund to overcome the health issues [2]. We argue to stop molecular biology and recombinant technology in every corner of this globe and perhaps one good centre in each big countries like USA, Canada, Germany, India, China, Brazil etc. is sufficient and that will reduce antibiotic use and artificial genetic recombination. G-20 Nations acted at Germany recently (July, 2017) in quite similar way but not exactly the same and we need more rigorous one Nation platform reducing unnecessary use of toxic chemicals in research [26]. Pesticides, paints, detergents should be biocompatible and user friendly as we saw poor farmers are using repeated pesticides in farmlands hoping good crops and now also are using bio-fertilizers (bacteria) as all nitrogen fixing bacteria have died in farmlands due to high dose of antibiotics and insecticides. In other words, stop this Earth from chemical toxicities [27]. Otherwise, MDR bacteria horror will be increasing with millions of such death in every country onwards 2050! I have surprised to see how the Ganga River water at Kolkata (West Bengal, India) has contaminated with MDR bacteria resistant to most common antibiotics including most deadly drugs like imipenem, amikacin, linezolid, lomofloxacin, vancomycin, cefotaxime, methicillin and ceftrioxane. MDR bacteria present in air dust particles and thus travel to any place with wind and fall in any place during rain. MDR bacteria has been detected in many asymptomatic animal and human with no detection of injury.

Figure 6: Gut microbiota populations (2x1012). Such bacteria are necessary for vitamin synthesis and many exmetabolites trigger IL6, IL22, IL26 and LPS production which send signal to brain for immunoregulation between intestinal cells and bacteria. Research indicated that imbalance in bacterial populations may facilitate modern disease epidemics like cancer, autisin, diabetes and cardiovascular diseases. Chronic inflammation leads to tissue destruction and complications. Chronic lowgrade inflammation is associated with obesity and metabolic dysfunction (insulin resistance). Increased LPS levels can stimulate eCB1 receptors, which activates the endocannabinoid and promotes adipogenesis [19].
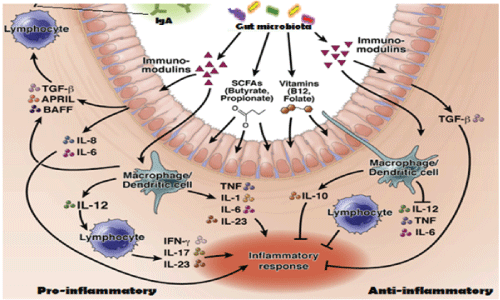
Figure 7: Symbiotic relation between gut bacteria and intestinal cells to produce inflammatory cytokines. Immunomodulins, vitamins and SCFAs modulate intestinal cells to produce pro-inflammatory cytokines (IL-6, IL-10, TGFβ, IL-23, IFN-γ) and anti-inflammatory cytokines (IL-10, TNF, IL-12, IL-17). Our hypothesis have suggested that such cytokines are also released by macrophages and dendritic cells inducing molecular changes in bacteria to create gene rearrangements and mdr gene creation to save gut microbiota [20,21].
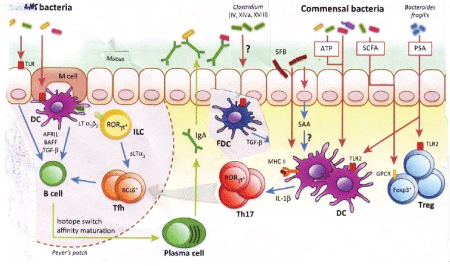
Figure 8: Adaptive immune-regulation by gut bacteria. Microbiota regulates intestinal immune responses primarily through the production of pathogen-associated molecular patterns (PAMPs) and metabolic byproducts. Microbiota stimulation leads to B cell switch to IgA, regulatory T cell induction, and T cell differentiation to Th17. Translocation of bacteria and bacterial products, immune activation and proinflammatory cytokine production likely cause gene rearrangement to create mdr genes in intestinal bacteria with stress induced by antibiotics [22,23].
| Multi-drug resistant (mdr) genes in bacterial conjugative large Plasmids | ||||
| Accession number | Size (kb) | Beta-lactamases, drug transporters, metal resistant and antibiotic inactivating enzymes were found | GenBank Year | MDR Pathogens |
| NC_018107 | 353 | aac3’-IId, aph2’, terA/C/F, Sul1, ANT3’’-Ia, dhfr, blaCTX-M-3, aacA4, blaSHV-12, aph3’’, blaTEM | 2017 | K. oxytoca |
| NC_022078 | 317 | MFS, merBC, cat, sul1, aac3’, cmr, tetA, tetG, ABC, blaKPC, blaCTX-M-24, blaVEB-3, aph3’-Ia, copB/C | 2017 | K. pneumoniae |
| LN555650 | 299 | terA/C/F/W/Y, blaAmpC, sul1, arsB, silA, strA, dhfr, catA1, blaACC-1, aadA1, aacA4, blaVIM-1 | 2015 | S. enterica |
| KM877269 | 249 | aad, floR, hph, aac6’/3’, blaOXA-1, catB, arr3, sul1 | 2015 | S. enterica |
| CP011634 | 227 | blaOXA, aad, blaTEM, merC, aad, sul1, aac, blaTEM | 2015 | K. oxytoca |
| HG530658 | 223 | terW, blaACC-1, strA, aadA2, aac3’, rcnA, pcoS | 2015 | E. coli |
| LN850163 | 167 | MFS, AAA tetA, cat, blaTEM, macAB, blaCTXm | 2015 | E. coli |
| KT185451 | 151 | blaTEM/CTXm/SHV12/KPC, merD, blaNDM1 | 2015 | K. pneumoniae |
| KF705205 | 134 | hph, strA, aac(3’)-IV, tetA, blaTEM-1 | 2015 | S. enterica |
| KP893385 | 137 | blaCTXm-65, blaKPC-2, blaSHV-12, blaTEM-1b | 2015 | K. pneumoniae |
| KC543497 | 501 | Ter2, blaOXA-10, MFS, blaTEM8, ble, catB8, aac | 2014 | P. aeruginosa |
| NC_012690 | 148 | floR, tetA, strB, sul2, blaAmpC, sul1, aph , blaTEM1, | 2014 | E. coli |
| HG941719 | 135 | bla TEM1 , aadA5, mphA, bla CTX , bla OXA , aac6, sulI, tetA | 2014 | E. coli |
| NC_020087 | 133 | aphA, hph, tetA, blaLAP2 , dhfrXII, ble, qnrS1 | 2014 | K. pneumoniae |
| NC_019375 | 180 | blaVIM, aacA7, dhfr, ANT3’, SHV-5, sul1, aph3’ | 2014 | P. stuartii |
| NC_022522 | 168 | blaCTX-M25, aacA4*, strB, strA, aadB, blaOXA21 | 2014 | S. enterica |
| NC_019121 | 166 | blaAmpC, sul2, tetA, floR, TniB, mcp, hygB, aph | 2014 | S. enterica |
| CP007558 | 272 | blaAmpC, ABC, sul1, blaTEM, aad, ble | 2014 | C. freundii |
| AP012055 | 250 | blaNDM , ccdA, ccdB, aadA2, catA1, qacA1 1 | 2013 | K. pneumoniae |
| AP012056 | 141 | Aac3/6, catB4, tetA, sul2, blaOXO/CTX/TEM, strB A / | 2013 | K. pneumoniae |
Table 2: Localization of many mdr genes in MDR conjugative plasmids of diverse pathogenic bacteria. GenBank search indicated as high as 5-15 mdr genes located in single large plasmid indicating how selective presser was mounted in bacteria to create mdr genes and its accumulations in single plasmids. Our data previously pinpointed the presence of large as well as small plasmids in MDR bacteria [12]. Bla genes lyses penicillin drugs, aac and aph genes acetylate and phosphorylate drugs respectively, and drug transporter like tet, mcr, macAB, mexAB (MFS, RND) kick out drug from bacterial cytoplasm. Such composite plasmids are created in acute stress of intestinal cells due to lack of vitamins during high dose antibiotics treatment on gut microbiota [2,12].
So we conclude: (i) Penicillin, cephalosporin and carbapenem drugs are highly eradicated gut microbiota and thus various isoforms of drug resistant beta-lactamases are generated; (ii) Tetracycline, streptomycin and sulfamethoxazole use are rampant and similarly tetA, tetB, tetC and tetM as well as strA/B and sul1/2/3 genes are very frequent in plasmids and chromosomes (Table 2); (iii) Acetyl/Phospho/Adenyl transferases (cat, aac, aad, aph genes) are also frequent in plasmids due to over exposure of chloramphenicol, aminoglycosides (erythromycin and azithromycin) and fluroquinolones (ciprofloxacin); (vi) many drug efflux genes accumulated, mutated and activated in plasmids but also in bacterial chromosome (Table 3); (v) thus any drug in over use will follow drug resistant (like, NDM-1, Mcr-1 genes) in human and animal because sensing mechanisms for gene rearrangement and mdr gene generation are protected from both sides i.e. bacteria and host (human and animal) without which both will be extinct.
| Chromosomal spread and localization of multi-drug resistant genes | ||||
| Names of the Bacteria | Chromosome acc. no./Size (bp) | mdr gene/% similarity | Position in bacterial chromosome/ Size (bp) | Copy no. |
| Escherichia coli BW25113 | CP009273/4631469 | acrAB/100% envCD /100% cmr/ 99% | 476132-482170/ 6038 3400624- 3411591/ 11k 878844-880412/1568 |
one three one |
| Klebsiella pneumoniae | NC_021232/5270770 | emrD/99% acrAB/82% | 29052-30607/955 4323257-4328383/5127 |
one two |
| Stenotrophomonas maltophilia (MDR) | AM743169/4851126 | mexAB/80% mexEF/79% aph/80% | 4177460-4178947/1487 1879272-1880782/1510 2143348-2144475/1127 |
two one |
| E. coli 0103:H2 (Toxin producing) | NC_013353/5449314 | acrAB/100% envCD/98% cmr/100% | 489265-495303/6038 4090536- 4094991/4455 968010-969578/1568 |
one one one |
| Salmonella enterica | CP007557/4685859 | acrAB/84% acrB/74% emrD/82% | 508579-514081/5507 3444035-3447110/3087 3874357-3875522/1165 |
two
one |
| Bacillus thuringiensis | NC_005957/5237682 | PBP/ n.d. MFS, ABC | 2101873-2103330/1457 n.d. |
one many |
| Acinetobacter baumannii TCDC-AB0715 (MDR) |
CP002522/4138388 | bla /99% OXA-23 mexD/66% aadA1/A4/99% |
2760564-2761566/1002 2169029-2169609/580 267616-266813/803 etc |
one one three |
| Bacillus subtilis | AP012496/4043042 | bmr/99% | 2326257-2327658/1401 | one |
| Citrobacter spp.S-77 | NZ_DF830265 | blaCMY-13/86% | 212590-216842/4252 | one |
| S. aureus OCL, MRSA | CP000046/2934567 | norA/94% Lactamase-B/ n.d. | 775486-778125/2639 73402-74726/424 |
one one |
| Klebsiella pneumoniae | FO834906/5438894 | acrAB/82% acrB/77% | 1458887-1463969/5127 42401264240699/573 |
three |
Table 3: Chromosomal spread of mdr genes. GenBank search (www.ncbi.nlm.nih.gov/blast) of whole genomes were indicated the presence of many mdr genes in bacterial chromosome to increase the gene dose for antibiotic destruction and to save intestinal bacteria. acrAB, mexAB, blaOXA-23, PBP2, mcr, blaCMY, norA, aph, bmr, aadA1, are few mdr genes shown here.
G-20 Nations and WHO action Plans are important to reduce antibiotic use but other actions may be important against MDR horror [11,13]: (i) Immediate action plans are needed to stop generation of complex antibiotics derivatives that create more pressure on bacterial metabolism which act as catalysis for new gene creation; (ii) Mandatory use of vitamin complexes is necessary and combination of synbiotics with antibiotics is must to save the human cells from crisis of metabolosome; (iii) Direct coenzymes like NADH, FADH2, THFA and PLP may help patients and will reduce the mdr gene generation and must be supplied to patients during each antibiotic therapy; (iv) MDR-bacteria with 5-15 mdr genes in MDR conjugative plasmids and chromosomes are very frequent now and phage therapy is a new frontier for medicine, (v) Complex biomolecules made by microbiota like colipan, linoleic acid, p-cresol etc. will be implement in diet of patients and such research will be accelerated to keep human in this Earth for long time and (6) Phytoantibiotics in combination with gene medicines (SiRNA, Ribozymes, CASPER-CAS and others) and drug nanocarriers will be the future effective treatment options (Figure 9).
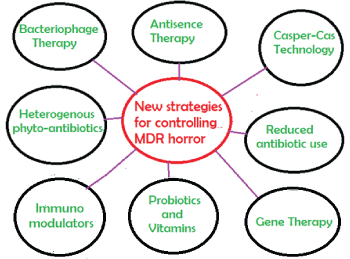
Figure 9: New strategies for controlling propagation of mdr genes in conjugative plasmids and chromosomes with rapid spread of superbugs. Such strategy must be adapted because use of antibiotics will be continued for sometime even more mdr genes will create and spread.
It is thus concluded that use of antibiotics without immediate probiotics or vitamin supplementation signalled to bacteria to generate mdr genes in household gut bacteria that will be thus able to continue synthesis of vitamins and coenzymes as well as other complex biomolecules absolutely needed at every point of human metabolosome [11, 28]. However, the signalling nature of intestinal cells to bacteria has not clearly understood. Active research is needed to know such signalling but antibiotic use also must be regulated. All points of research indicated that drug resistant bugs are more prominent at the site of industry that synthesis it and such contamination of drugs in many rivers is shocking [29-31]. Colistin is one of the last resort superbug drugs after imipenem (which lost in 2009 due to blaNDM-1 discovery & spread) but recently colistin was taken out from drug regime as Mcr-1 gene discovered in 2016 and its spread continue [32]. Also multiple dose of drug use by patients indeed have helped to generate MDR-bacteria suggesting that human intestinal sites are the place of bacterial gene rearrangement and mdr gene synthesis [11]. In truth MDR-plasmids are also increased the gene doses of integrases, transposes, recombinases and many ISelements. Phage therapy, gene medicines and nano-drug carriers may be alternative approaches to combat MDR horror [32]. Levamisole, Situximab, Prednisone and betaglycan immunomodulators have shown to induce suppressed immunosystem clearing mdr-TB by antibody production, T-cell activation and increased phaagocytosis and chetotaxis of macrophages [30]. Jemson KC et al. (2015) and others have disclosed the rapid use of bacteriophages in the treatment of MDR infections [31- 36]. Gene therapy using interleukin and cytokine genes in expression vectors with fullerenes or DNA-based nanocarriers are centre stage of development to cure MDR infections [37-39]. I hope MDR mechanisms caused very serious genetic effects in bacteria and huge funding must be allotted for basic research to understand the mdr gene creation and its spread into conjugative plasmid and chromosome.
I thank Dr J. B. Medda of OAER for financial support and Dr. Bidyut Bondhopadhyay for help during the study.
Download Provisional PDF Here
Article Type: Research Article
Citation: Chakraborty AK (2017) MDR Genes are Created and Transmitted in Plasmids and Chromosomes to Keep Normal Intestinal Microbiota Alive against High Dose Antibiotics- A Hypothesis. J Mol Med Clin Appl 2(1): doi http://dx.doi. org/10.16966/2575-0305.109
Copyright: © 2017 Chakraborty AK, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Publication history:
All Sci Forschen Journals are Open Access