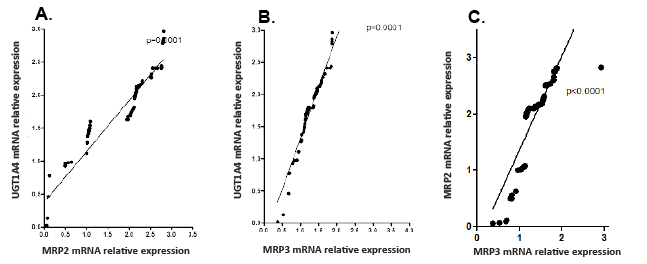
Figure 1: Correlation between mRNA relative expressions of A) UGT1A4 and MRP2, B) UGT1A4 and MRP3 and C) MRP2 and mRP3 in liver tissue samples.
Vineetha Koroth Edavana1 Rosalind B Penney2 Aiwei Yao- Borengasser1 Athena Starlard-Davenport1 Ishwori B Dhakal1 Susan Kadlubar1*
1Division of Medical Genetics, College of Medicine, University of Arkansas for Medical Sciences, Little Rock, USA*Corresponding author: Susan Kadlubar, PhD, University of Arkansas for Medical Sciences, 4301 W. Markham, #580, Little Rock, AR 72205, USA, E-mail: sakadlubar@uams.edu
Aritcle Type: Research Article
Citation: Edavana VK, Penney RB, Yao-Borengasser A, Starlard-Davenport A, Dhakal IB, et al. (2015) Effect of MRP2 and MRP3 Polymorphisms on Anastrozole Glucuronidation and MRP2 and MRP3 Gene Expression in Normal Liver Samples. Int J Cancer Res Mol Mech 1(3): http://dx.doi.org/10.16966/2381- 3318.112
Copyright: © 2015 Edavana VK, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Publication history:
Anastrozole is an aromatase inhibitor (AI) used as adjuvant therapy for breast cancer. Anastrozole is subject to direct glucuronidation catalyzed by UDP-glucuronosyltransferase1A4 (UGT1A4). Interindividual variability in anastrozole glucuronidation may be affected by UGT1A4 SNPs. Interplay between drug metabolizing genes such as UGT1A4 and transporter genes may also be affected by genetic variability. Thus, we hypothesize that genetic variability in MRPs could influence anastrozole glucuronidation.
The correlation between UGT1A4 and MRP2 or MRP3 transporter gene expressions and the correlation between MRP2 or MRP3 mRNA and anastrozole glucuronidation were analyzed in normal human liver samples. MRP2 and MRP3 mRNA levels were significantly correlated with UGT1A4 mRNA, with anastrozole glucuronidation and with each other (p<0.05). The data also demonstrated that MRP2 SNPs are positively correlated with MRP2 mRNA expression, while there was no association between MRP3 SNPs from this study and MRP3 expression. Significant correlations (p<0.05) between certain MRP2 SNPs (3972C>T, 2366C>T and -24C>T) and anastrozole glucuronidation were observed. There were no observed correlations between MRP3 SNPs and anastrozole glucuronidation.
MRP2 polymorphisms have been identified as playing a role in the disposition of other drugs, and the data presented here indicate for the first time that MRP2 SNPs could influence anastrozole metabolism and contribute to interindividual variation in treatment responses.
MRP2 Polymorphisms; MRP3 Polymorphisms; Anastrozole Glucuronidation; MRP2 gene expression; MRP3 gene expression
Breast cancer is the most frequently diagnosed cancer in women and the second most common cause of cancer-related death in women. In developed countries, around 75% of all breast cancers occur in postmenopausal women, about 80% of which are hormone-receptor positive [1]. Until recently, tamoxifen (TAM) has been the endocrine treatment of choice for postmenopausal women with early-onset hormone receptor-positive breast cancer. In the past decade, a number of aromatase inhibitors (AIs) have been developed as an alternate approach to TAM for the treatment of estrogen receptor-positive breast cancer. The current third-generation AIs (anastrozole, exemestane and letrozole) are highly specific to the aromatase enzyme and have fewer side effects than previous generations of AIs. Evidence from several clinical trials indicates that anastrozole may be superior to TAM as a first-line therapy for postmenopausal women with metastatic breast cancer. Results from at least eight major clinical trials indicate that anastrozole alone is associated with longer disease-free survival than therapy with TAM alone [2,3], which supports the use of anastrozole as a first-line therapy.
Although anastrozole has demonstrated some superiority relative to TAM, many patients still experience a recurrence of breast cancer after anastrozole therapy. In addition, there is also substantial interindividual variability with respect to tolerability. For example, musculoskeletal complaints can be so severe for some anastrozole patients that they withdraw from therapy [4]. This variability is consistent with differences among patients demonstrated in drug pharmacokinetics and/or pharmacodynamics studies and is potentially driven by host genetic variability [5].
Anastrozole belongs to the non-steroidal triazole-derivative group of AIs and is predominantly metabolized by Phase I oxidation followed by Phase II reactions, including glucuronidation. Anastrozole is also subject to direct glucuronidation catalyzed by UDP-glucuronosyltransferase1A4 (UGT1A4). The potential impact of UGT1A4 SNPs on anastrozole glucuronidation has been reported before [6-10]. UGT1A4 enzymes are localized with known UGT1A4 transport systems (such as MRPs), and both are induced by the same compounds, suggesting a correlated action [11]. Certain xenobiotics induce genes that encode transporter proteins. For example treatment of Caco-2 cells with the polyphenolic antioxidants quercetin and t-butylhydroquinone increased MRP2 expression. Interplay between transporters and drug-metabolizing enzymes have been postulated to have a major role in determining a drug’s absorption and disposition [12-14].
Interindividual variability of drug response is a widely recognized determinant of drug toxicities, especially for those drugs with narrow therapeutic windows. Glucuronidation activities in different human tissues have been shown to exhibit a high degree of variation [15-20,7]. Genetic polymorphisms are a major cause of such variability, often resulting in altered pharmacokinetics and subsequent pharmacological and toxicological effects of drugs. For example, single nucleotide polymorphisms (SNPs) of MRP2 contribute to interindividual variability in methotrexate, irinotecan and SN-38 drug disposition and, ultimately, in drug response [21].
Previous studies from this lab have reported that UGT1A4 mRNA, MRP1 mRNA, MRP2 mRNA and MRP3 mRNA are coordinately induced by fulvestrant in MCF7 and HepG2 cell lines. This upregulation of UGT1A4, MRP1, MRP2 and MRP3 mRNA was positively correlated with anastrozole glucuronidation [15]. These data demonstrate that the metabolizing gene UGT1A4 and transporter genes MRP1, MRP2 and MRP3 may play a role in drug disposition. Other studies from this lab observed a significant correlation of UGT1A4 expression levels with interindividual variability of anastrozole glucuronidation [15]. Small variations in the UGT1A4 gene promoter region altered constitutive expression of UGT1A4, and these variations had an effect on anastrozole glucuronidation. The clinical application of pharmacogenomics in cancer treatment with anastrozole will therefore require more detailed information concerning the functional effects of genetic variants in drug metabolizing enzymes and drug transporters.
To date, the potential impact of MRP SNPs on anastrozole glucuronidation has not been explored. To address this, we examined the correlation between glucuronidation of anastrozole in human liver microsomes and genetic variants of MRP2 and MRP3 (MRP1 was not explored as it is not expressed in the liver). These factors, if understood, would offer the potential for individualizing treatment and ensuring that patients receive optimal therapy.
Anastrozole was obtained from Toronto Research Chemicals Inc. (Toronto, Canada). Alamethicin, magnesium chloride, Tris-HCl and UDP glucuronic acid (UDPGA) were purchased from Sigma-Aldrich (St Louis, MO). Recombinant UGT1A4 was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). All other reagents were of HPLC grade or of the highest grade commercially available.
The HLMs used for this study have been described previously [22]. Briefly, human liver specimens (n=102) were obtained from the Cooperative Human Tissue Network (CHTN). All liver specimens were from Caucasian donors ranging in age from 26 to 102 years, with 54 male, 42 female and 6 unknown. African Americans were excluded from this study due to low numbers that precluded racial comparisons. All liver specimens were snap-frozen upon harvest and were confirmed as histologically normal tissue by CHTN. Tissue specimens that exhibited abnormalities were excluded from this study. Microsomes were prepared from human liver tissue as previously described [23]. Briefly, liver cytosol fractions were prepared at 4°C by differential centrifugation after homogenization in 4 vol of a solution containing 0.25 M sucrose, 50 mM Tris–HCl, pH 7.8, 0.5 mM EDTA, 20 µM butylated hydroxyl toluene and 0.1 mM dithio threitol. Cytosol fraction was removed, and pellets were resuspended in solution containing 0.25 M sucrose, 10 mM Tris–HCl, pH 7.8, 0.1 mM EDTA, 20 mM butylated hydroxytoluene and 20% glycerol. Samples were stored frozen at -80°C until assayed. Microsomal protein levels were determined using the Bradford method [24] with bovine serum albumin as a standard.
Total RNA was isolated using TRIzol (Invitrogen, Carlsbad, CA) and was used as a template for cDNA synthesis with Superscript II (Invitrogen, Grand Island, NY). Quantitative RT-PCR was performed using an ABI 7900HT Fast Real Time PCR System and SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA). Gene specific primers, annealing temperature and cycle numbers for UGT1A4, MRP2 and MRP3 have been described previously [15,25,26]. The dissociation curves for each reaction were examined to ensure amplification of a single PCR product in the reaction. The -fold change in mRNA levels was determined after normalizing the gene expression levels to those of β-actin.
The glucuronidation of anastrozole was measured in pooled HLMs, recombinant UGT1A4 and microsomes from 102 individual human livers as described previously [15]. Briefly, metabolite quantitation was performed using an Aquity ultra-performance liquid chromatography (Waters, Milford, MA) interfaced source to an TSQ-Quantum Ultra triple quadrupole (ThemoScientific, SanJose, CA) with use of a heat-assisted electrospray ionization source. An Aquity ultra-performance liquid chromatography bridged ethane-silicon hybrid C18 column was operated with a linear gradient from 5% acetonitrile/0.1% formic acid to 80% acetonitrile/0.1% formic acid in 5 minutes at 0.4 ml/min. Anastrozole and anastrozole glucuronide were acquired by monitoring the ion transition of m/z (mass-to-charge ratio) 294.0 to 225.2 and m/z 470.2 to 225.2, respectively. Because of the absence of authentic standards for anastrozole glucuronide, quantification of the glucuronide was accomplished using a standard curve for anastrozole. Enzymatic assays were performed according to standard procedure [7,5].
Genotyping for MRP SNPs was performed in 102 samples. Genotyping for MRP2 and MRP3 sequences were performed as described previously [19,7]. All amplification was performed using 1U of Taq Polymerase. Sequencing was performed using an ABI Prism 3700 DNA analyzer (Applied Biosystems).
Apparent kinetic parameters (Km and Vmax) for anastrozole glucuronidation were estimated by fitting apparent formation rates of metabolite vs. anastrozole concentrations to a single- or two-site Michaelis-Menten enzyme kinetic equation using nonlinear regression analysis software (GraphPad Prism Software Inc, Version 5, San Diego, CA) or the Enzyme Kinetics Module of SigmaPlot (Systat Software, Inc., San Jose, CA).
Both parametric and non-parametric tests were performed to examine the correlation of anastrozole glucuronidation with UGT1A4 mRNA, anastrozole glucuronidation and MRP SNPs. Also, UGT1A4 mRNA was correlated with MRP SNPs. In parametric one-way ANOVA, nonGaussian distributed variables were log transformed and analysis was implemented using ‘PROC GLM’. Non-parametric one-way ANOVA was executed using ‘PROC NPAR1WAY’. A p<0.05 (2-sided) was considered to be statistically significant and all analyses were performed using SAS software (version 9.2, Statistical Analysis Systems, Cary, NC).
MRP SNPs that appeared significantly associated (at α=0.05) with expression were used to construct haplotypes. PHASE program was used to predict the population frequencies and “best pair” of haplotypes for each individual as previously described [27]. Briefly, this program provides a “best pair” of haplotypes that most likely explains the genotypes of that subject with the corresponding probability of having that particular haplotype for each individual. Alternative pairs of haplotypes are assigned in the cases of uncertain haplotypes. PHASE output was translated into one variable for each haplotype with three possible values (0-, 1- or 2-copy) through the use of SAS version 9.3. Association of each haplotype with gene expression was examined again using nonparametric analysis of variance. p<0.05 (2-sided) was considered to be statistically significant.
To demonstrate co-regulation of UGT1A4 and MRPs, human liver samples were used. Correlation analysis was performed between UGT1A4 mRNA and MRP2 mRNA and UGT1A4 mRNA and MRP3 mRNA. UGT1A4, MRP2 and MRP3 mRNA expression levels were estimated using RT-PCR and correlation analyses were performed in 102 normal liver samples. Correlation studies demonstrated MRP2 expression levels were significantly correlated with UGT1A4 (r=0.8, p<0.001; Figure 1A). MRP3 expression levels were also significantly correlated with UGT1A4 mRNA expression levels (r=0.9, p<0.0001; Figure 1B). Correlation analysis was also performed between MRP2 mRNA and MRP3 mRNA, with relative expression demonstrating correlation (r=0.86, p<0.0001; Figure 1C).
Interindividual variability in anastrozole glucuronidation was evaluated by LC-MS/MS in liver microsomes collected from the same 102 individuals. Interindividual variability of anastrozole glucuronidation ranged from 0.0005-0.009 pmol/mg/min protein. MRP2 and MRP3 mRNA levels were normalized against β-actin. MRP2 and MRP3 mRNA expression levels were then correlated with anastrozole glucuronidation. MRP2 and MRP3 mRNA expression levels were significantly correlated with glucuronidated anastrozole levels (p<0.001) (Figures 2A and 2B), but not with the parental anastrozole compound (p>0.5, data not shown).This corresponds with the results from our previous cell line experiments.
To determine the contribution of MRP2 and MRP3 SNPs to anastrozole glucuronidation, dideoxy sequencing was performed on genomic DNA from liver specimens of 102 human subjects. The functional MRP2 SNPs (3972C>T, 2366C>T, -24C>T, 4348 C>A, 1249G>A, 4430C>T, 4544 G>A and 3563 T>A) reported to have contribution in MRP2 activity were genotyped (Table 1A). Sequencing analysis of MRP2 revealed three MRP2 SNPs (3972C>T, 2366C>T and -24C>T) were significantly correlated with anastrozole glucuronidation (p<0.001; Figures 3A-3C). Microsomes from samples with variant T allele at position 3972 exhibited significantly increased glucuronidation activity toward anastrozole compared to microsomes from samples variant T alleles for the C<T and C<T at positions 2366 and -24 respectively (Figure 3D).
Also, 3972C>T (p=0.001), 2366C>T (p=0.001) and -24C>T (p=0.001) were significantly correlated with MRP2 mRNA expression levels (Figures 4A-4C).

Figure 1: Correlation between mRNA relative expressions of A) UGT1A4 and MRP2, B) UGT1A4 and MRP3 and C) MRP2 and mRP3 in liver tissue samples.
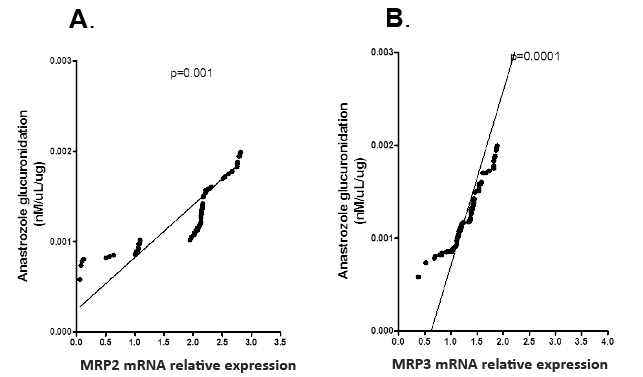
Figure 2: Correlation between anastrozole glucuronidation and A) MRP2 mRNA relative expression and B) MRP3 mRNA relative expression in liver tissue samples.
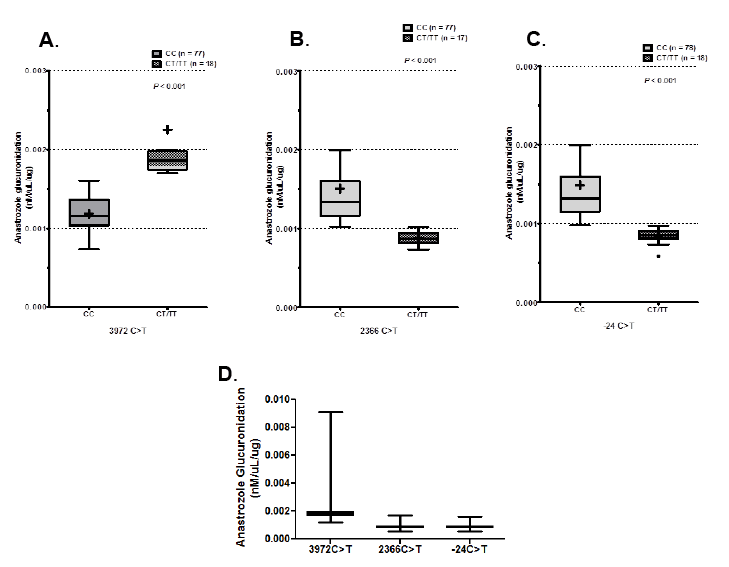
Figure 3: Samples were sequenced and genotypes correlated with anastrozole glucuronidation. Significant differences in glucuronidation were identified between the homozygous common genotype and hetero- or homozygous variant genotypes in A) 3972C>T, B) 2366C>T and C) -24C>T. D) When anastrozole glucuronidation was compared between variant genotypes of these SNPs, variant T allele at 3972C>T had higher glucuronidation than the variants at 2366C>T or -24C>T.
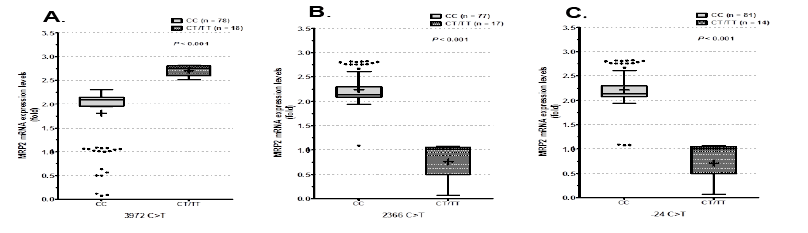
Figure 4: Genotypes within 3972C>T, 2366C>T and -24C>T were correlated with MRP2 mRNA expression. Significant differences in MRP2 mRNA expression were identified between the homozygous common genotype and hetero- or homozygous variant genotypes in A) 3972C>T, B) 2366C>T and C) -24C>T.
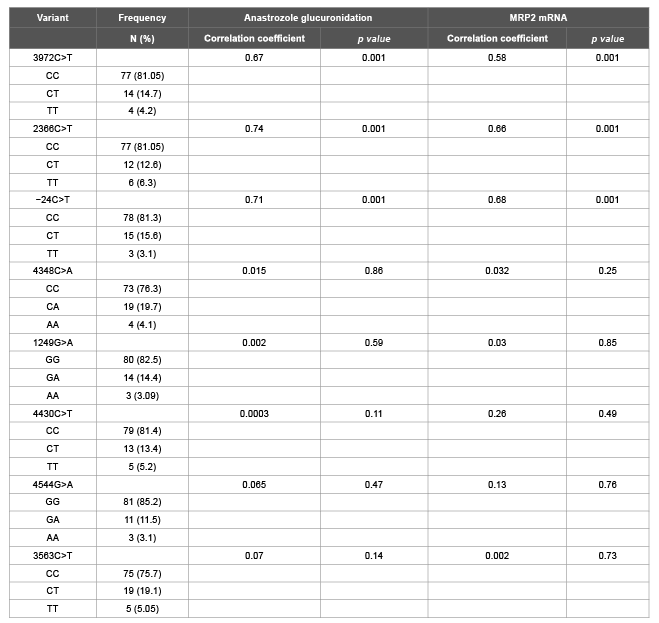
Table 1A: Analysis of anastrozole glucuronidation and MRP2 mRNA expression levels by MRP2 variants
Previously reported MRP3 SNPs (-211C<T, -1767 C>T, 202C>T, 1037C>T, 1537C>A, and 3890G>A) were also genotyped in these liver samples. These MRP3 SNPs did not show any significant correlation with anastrozole glucuronidation or with MRP3 expression (Table 1B).
The MRP2 3972C>T, 2366C>T, -24C>T haplotype TCC was significantly associated with anastrozole glucuronidation (p=0.0001, frequency 0.198; Table 2). No other haplotypes were significantly associated with anastrozole glucuronidation.
Aromatase inhibitors have been approved as a first-line treatment option for advanced hormone-dependent breast cancer in postmenopausal patients. However, emerging data suggest that high interindividual variability can lead to both beneficial and adverse effects of these drugs [4].
Anastrozole, a member of the nonsteroidal triazole-derivative group of aromatase inhibitors, is metabolized (activated) and cleared mainly through N-dealkylation, but also through hydroxylation via the CYP3A4 enzyme [28,29] and through glucuronidation via the UGT1A4 enzyme [5].
Interplay between transporters and drug-metabolizing enzymes have been postulated to have a major role in determining a drug’s absorption and disposition [12-14]. Previous studies from our group report that the metabolizing gene UGT1A4 mRNA and the transporter genes MRP1, MRP2 and MRP3 mRNA were coordinately induced by fulvestrant in MCF7 and HepG2 cell lines. These upregulations are positively correlated with anastrozole glucuronidation in the cell lines. This lab has also demonstrated a significant correlation of UGT1A4 expression levels with interindividual variability of anastrozole glucuronidation in liver microsomes [15]. These data demonstrate that the metabolizing gene UGT1A4 and transporter genes MRP1, MRP2 and MRP3 may play a role in drug disposition. Genetic variants that lead to constitutive expression and alter the ability to respond to physiological inducers contribute to interindividual variability in glucuronidation capacity. This could be caused by several factors, one of which could be the presence of functional genetic variants. The clinical application of pharmacogenomics in cancer treatment will therefore require more detailed information concerning the functional effects of genetic variants in drug metabolizing enzymes and drug transporters.
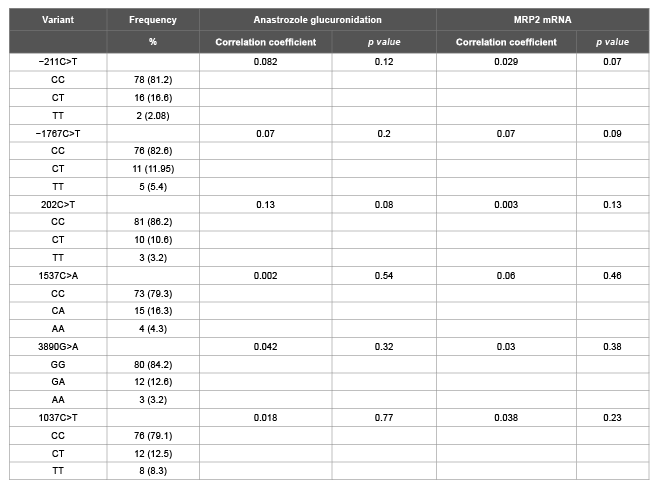
Table 1B: Analysis of anastrozole glucuronidation and MRP3 mRNA expression levels by MRP3 variants
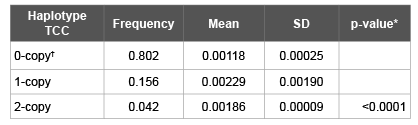
Table 2: Haplotypes created based on results from microsomal anastrozole glucuronidation as noted in Methods.
SD: Standard Deviation
*Non-parametric ANOVA (Kruskal-Wallis test)
†0-copy=no copies of haplotype, 1-copy=one copy of haplotype,
2-copy=two copies of haplotype
Drug absorption and disposition may be determined by the interactions between transporters and drug-metabolizing enzymes. The potential impact of UGT1A4 SNPs on anastrozole glucuronidation has been reported [6-10]. To date, the potential impact of MRP SNPs on anastrozole glucuronidation has not been explored. To address this issue, we examined the glucuronidation of anastrozole in human liver microsomes and analyzed the correlation of genetic variants of MRP2 and MRP3 on anastrozole glucuronidation. MRP1 SNPs were not explored because MRP1 is not expressed in the liver.
In previous studies, a marked interindividual variability has been observed in anastrozole glucuronidation and UGT1A4 mRNA expression in liver microsomes. In this study, a similar trend in interindividual variability was observed in MRP2 mRNA expression and MRP3 mRNA expression levels in liver microsomes. MRP2 mRNA correlated with UGT1A4 mRNA expression and MRP3 mRNA correlated with UGT1A4 expression levels in these liver samples (p<0.001). It has been reported that UGT1A family members and MRP family members have both been upregulated in the presence of a drug [30,31], but to our knowledge this is the first demonstration of the co-regulation of metabolizing genes and transporter genes in liver microsomes. These results also support previously published data in cell lines [15].
There was a significant correlation between MRP2 mRNA expression and anastrozole glucuronidation and MRP3 mRNA expression and anastrozole glucuronidation (p<0.001). However, there was no correlation with MRP2 or MRP3 expression levels and the anastrozole parent compound (data not shown). This indicates that MRP2 and MRP3 maybe co-regulated with only glucuronidated anastrozole and not the parental compound. We observed similar results in our previous MCF7 and HepG2 cell lines study [15].
Glucuronidation activities in human tissues have exhibited a high degree of interindividual variation in human liver samples which can be partially explained by UGT1A4 [16,17,32,33]. Single nucleotide polymorphisms (SNPs) may influence the genetic role in variability. Previous data suggest that genetic variants in UGT1A4 could impact anastrozole pharmacogenomics. The presence of SNPs within the promoter and coding regions of the UGT1A4 gene can lead to quantitative or qualitative alterations of specific catalytic activities between individuals [15,18,20,7]. This study proposes that the interindividual variability could also be affected by variation in MRP2 and MRP3. In this study the association between MRP2 and MRP3 functional promoter and coding SNPs and variability in anastrozole glucuronidation was explored. MRP2 functional SNPs were correlated with both gene expression and anastrozole glucuronidation. In liver microsomes from individuals with variant ‘T’ allele (homo or hetero) at MRP2 positions 2366 and -24, there was a positively correlated decrease in MRP2 mRNA expression. There was also a decrease in anastrozole glucuronidation. On the other hand, microsomes from individuals with variant ‘T’ allele (homo or hetero) at position 3972 exhibited significantly higher MRP2 gene expression, as well as higher anastrozole glucuronidation activity. Our results are consistent with previous reports of the functional effect of 3972C>T, 2366C>T and -24C>T SNPs with MRP2 expression levels [34,21,35]. The -24 C>T SNP in the 5’ untranslated region of MRP has been associated with altered pharmacokinetics for various drugs like methotrexate, irinotecan and irinotecan’s active metabolite SN-38 [36-38]. It could be that these results are partially due to the coordinate regulation of UGT1A family members and MRP family members. However, previous reports analyzed the mechanism of the SNP variant 2366C>T and found that 2366C>T decreased the expression (40%) of all MRP2 and demonstrated a 40% decrease in the transport of MRP2 substrates. To our knowledge, the current study describes for the first time the association of MRP2 SNPs and anastrozole glucuronidation.
To our knowledge, only one MRP3 promoter SNP (-211 C>T) has been reported to have functional significance. This MRP3 SNP was selected for analysis in this study, as well as five chosen from NCBI. There was no observed correlation between MRP3 SNPs and MRP3 expression or with anastrozole glucuronidation. There were no demonstrated MRP3 effects, though there is correlation between gene expression and activity reported by others. Lang et al. [39] reported that the -211C>T promoter polymorphism appears to be associated with altered hepatic MRP3 mRNA expression, but this effect was not observed in the current study. It is possible that the functional SNP chosen does not target anastrozole glucuronidation, and that the other SNPs chosen did not include functional SNPs. Future studies will include a wider array of MRP3 SNPs. Further detailed screening of other functional alleles which have an impact on MRP3 expression levels need to be done to reveal the contribution of MRP3 in anastrozole glucuronidation.
Haplotypes were developed with the MRP2 SNPs found to be significant to anastrozole glucuronidation. Haplotype/activity analysis demonstrated that MRP2 3972C>T, 2366C>T, -24C>T haplotype TCC was significantly associated with anastrozole glucuronidation if the sample had either 1 or 2 copies. There is a linkage reported between 3972C>T and 2366C>T SNPs with -24C>T SNP. Ito et al., 2001 and Itoda et al. [35] reported a strong association between -24C>T and 3972C>T which has a role in the regulation of reducing MRP2 expression and MRP2 promoter activities, as well as efflux activity [40,35]. The 3972C>T SNP in exon 28 is synonymous and is not expected to be functional. However, the linkage between this SNP and -24C>T SNP has explained its effect on MRP expression levels and promoter activities.
In conclusion, the results from the present study suggest that interindividual variability of anastrozole glucuronidation is correlated with MRP2 SNPs and gene expression. Further studies examining MRP2 SNPs and anastrozole pharmacokinetics are warranted.
The clinical relevance of MRP2 polymorphisms in the disposition of drugs such as pravastatin [41] and anticancer drugs such as irinotecan [42] and tamoxifen [43] is well established. These SNPs may also prove to be of toxicological and clinical relevance since MRP2 is an important component of the detoxification of environmental carcinogens, e.g., 2-amino-1-methyl-6-phenylimidazo [4,5- b]pyridine (PhIP) [44] and arsenite glutathione complexes [45]. These data indicate that closer evaluation of the role of these MRP2 SNPs in haplotype analyses or genome wide association studies of adverse drug reactions and/or disease is warranted, as is careful monitoring of patients with the relatively frequent MRP2 SNPs.
This work was supported by the National Institutes of Health National Cancer Institute [Grant R01CA118981].
The authors declare they have no conflicts of interest to disclose.
VKE, RBP and SK participated in research design, VKE, AY-B and AS-D conducted experiments, VKE, IBD, AY-B and RBP have performed data analysis and VKE, RBP, AY-B and SK wrote or contributed to the writing of the manuscript.
Download Provisional pdf here
All Sci Forschen Journals are Open Access