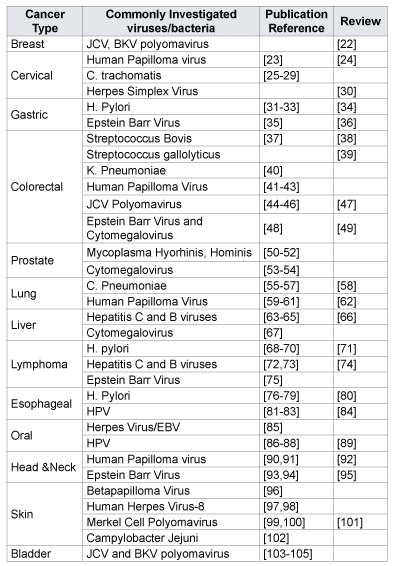
Table 1: Cancer types and commonly investigated viruses and bacteria with selected references.
Microbiome Analysis: Trends in Cancer Epidemiology, Challenges and Opportunities
Elizabeth F Hebert, Rao L Divi, Mukesh Verma*Epidemiology and Genomics Research Program, Division of Cancer Control and Population Sciences, National Cancer Institute, Rockville, MD 20850, USA
*Corresponding author:Mukesh Verma, Ph.D. Chief, Methods and Technologies Branch,
Program Director, Epidemiology and Genomics Research Program,Division of Cancer Control
and Population Sciences, National Cancer Institute (NCI), National Institutes of Health (NIH),
Suite# 4E102. 9609 Medical Center Drive, Rockville, MD 20850, USA, Phone (240) 276-6889 ;
Fax (240) 276-7909 ;
E-mail: vermam@mail.nih.gov
Article Type: Review Article
Citation: Hebert E, Rao LD, Verma M (2015) Microbiome Analysis: Trends in Cancer Epidemiology, Challenges and Opportunities. Int J Cancer Res Mol Mech, Volume1.1: http://dx.doi. org/10.16966/2381-3318.101
Copyright: © 2015 Verma M. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Publication history:
Analysis of microbiome sampled from a given cancer site can yield information that may serve as a prognostic of exposure, risk, disease progression, and treatment response. We reviewed available published literature, and grants funded by the National Cancer Institute’s Division of Cancer Control and Population Sciences to identify trends and areas for future research. The incorporation of microbiome analysis in epidemiologic studies of cancer is providing some promising insights into risk stratification. However, our analysis identified several knowledge gaps in this emerging field: 1. Limited evidence for the stability of different biospecimen types over time and at given temperatures for microbiome analysis, 2. Analysis software that reliably standardizes sequencing data from different platforms and corrects for biases against rare or unrepresented taxa, 3. Harmonization of methods used for microbiome analysis across research centers. 4. Surrogate markers that will be useful for monitoring disease progression from the time of infection to cancer development, 5. Time-varying microbiome in the natural history of different cancers in order to identify key microbiota or shifts in community structure, and 6. Determining whether the microbiota cause or effect cancer risk and outcomes. If these knowledge gaps are addressed, microbiome analysis is likely to provide the cancer field with new approaches for early diagnosis and help develop more effective preventative measures.
Microbiome; Microbiota; Cancer; Epidemiology
The microbiota that colonize various anatomical sites throughout the human body contribute to our overall health through physiologic and metabolic processes necessary for survival. Microbiome is the associated genome of the microbiota and it codes for the necessary processes that are not encoded by the human genome [1]. From birth, commensal microbiota prime our immune system to prepare for the millions of immunologic insults we encounter throughout our lives [2,3]. Microbiota acquired early in life are responsible for these functions and remain relatively stable for most of our lives [4,5]. The microbiota is also dynamic; the abundance or functions of certain species within a community can shift or change in response to typical exposures such as: infection, antibiotics, and/or diet. In some cases these shifts propagate disbyosis and dysfunction of microbiota which contribute to disease [6].
It has been demonstrated that disbyosis of the gut microbiota can lead to a chronic inflammatory response and an environment that promotes cancer progression [7,8]. Several studies have combined functional genome analysis of 16S rRNA regions and metabolite analysis of stool samples from advanced colorectal adenoma patients to analyze changes in the microbiome. They identified decreases in butyrate-producing bacteria in cancer associated microbiomes, as well as increased concentrations of bile salts in stool samples from diseased compared to healthy controls [9-13]. It has also been shown that the gastrointestinal microbiome modulates prostate cancer risk through the metabolism of plant phenols, calcium, and choline compounds [14]. Disbyosis of the oral microbiome has been investigated in association with pancreatic cancer [15-17] and oral cancer [18,19].
Analysis of the microbiome over time can yield information that may serve as a diagnostic of: microbial exposure, cancer risk, incidence, and progression, as well as treatment response. The study of the microbiome in various cancer cohorts may also identify modifiable risk factors and mechanisms of carcinogenesis, which may inform preventative measures and early diagnosis.
In order to better understand the potential applicability of microbiome analysis in cancer epidemiology, a review of the published literature, and the grants funded through the National Cancer Institute’s Division of Cancer Control and Population Sciences, was conducted and trends in cancer site, sampling, methods and technologies used were identified. The advantage of comparing published literature and funded grants is significant because the précis generates novel ideas and hypothesis for further research and new approaches in disease intervention and treatment. This review also allowed for the identification of knowledge gaps in the current literature and opportunities for future research.
Materials and Methods
Criteria and terms used for identifying microbiome and cancer epidemiology grants and publications: search strategy and analysis
Microbiome grants funded by the National Cancer Institutes’ Division of Cancer Control and Population Sciences from January 1st, 2009 to December 31st, 2013 were included in the portfolio analysis using the terms ‘Microbiome’ and ‘Cancer’. This portfolio was created using the Portfolio Management Application software version 16.0. From this data search, a total of 40 grants were selected for the portfolio analysis. Prior to data collection, the selected grants were analyzed with the following inclusion criteria: (i) the focus of the project was cancer; (ii) the project included at least 100 human cancer cases; and (iii) has a minimum of one microbiome-related specific aim. Animal model studies and in vitro analyses were excluded from final analysis and data collection of the 40 originally identified, 24 grants were included in the final analysis based on these criteria.
In addition to the grant portfolio analysis, a search of the published literature on the microbiome and cancer epidemiology available on PubMed from January 1st, 2009 to December 31st, 2013 was completed using the following search criteria: (“microbiota”[MeSH] OR “bacteria”[MeSH] OR “viruses”[MeSH]) AND (“infection”[All Fields] OR “dysbiosis”[All Fields]) AND (“neoplasms”[MeSH] OR “neoplasms”[All Fields] OR “cancer”[All Fields]) AND (“epidemiology”[subheading] OR “epidemiology”[All Fields] OR “epidemiology”[MeSH]) AND (“humans”[All fields] OR “humans”[MeSH]) AND (“2009/01/01”[PDAT] : “2013/12/31”[PDAT]) NOT “review”[ptyp] NOT “meta-analysis”[ptyp] NOT “in vitro”[All fields]. Applying the same inclusion criteria used for the grants, while also excluding meta-analyses and reviews, 284 publications were identified for further analysis. Information from these grants and publications were collected and coded for: cancer type/site, population demographics, study design, microbiome measure (i.e. strains/viruses investigated or community structure), technology used, and sample types collected.
Results
Figure 1 shows the cancer sites investigated with associated microbiomes in NCI-funded grants and the published literature. Cervical is the most studied cancer site in the publications followed by gastrointestinal; whereas, colorectal and liver are the most studied cancer sites in NCIfunded grants, respectively. The majority of the publications on cervical cancer assessed HPV infection and cancer. Similarly, publications on gastrointestinal cancer assessed for H. pylori infection. Table 1 shows commonly investigated infectious agents and associated cancer sites. Given our established inclusion criteria, very few publications included in this analysis used true microbiome analysis and instead focused on identifying known or commonly associated carcinogenic infectious agents. Trends in the NCI-funded grants are demonstrating a shift to true microbiome analysis in cancer epidemiology, by assessing the overall changes in community structure or function. It is also important to note that NCI-funded grants are using methodology that is up to date with the present practices for microbiome analysis and show the current trend.
The types of samples collected for microbiome analysis in the published literature and NCI-funded grants are shown in Figure 2. In the grants, stool samples and serum samples were the most commonly collected biospecimens; whereas, tumor tissue and cervical samples were the most commonly collected biospecimens in the publications. Trends in collected biospecimen types reflect the commonly studied cancer sites with associated microbiomes.
The trends in the methods and technologies used for microbiome analysis are shown in Figure 3. In the NCI-funded grants, targeted 16S sequencing followed by PCR based methods for confirmation is the most commonly used method for microbiome analysis. In comparison, ELISA and PCR based methods are the most commonly used methods for analysis within the publications. Though targeted 16S sequencing is the most common method of analysis for microbiome analysis, most studies that utilized targeted 16S sequencing typically included sample sizes of less than 100 cancer cases. Based on our criteria for analysis of the grants and publications, we had to exclude these small scale studies from our analysis. However, this trend toward targeted sequencing using 16S rRNA primers for microbiome analysis in epidemiologic studies is demonstrated in its use in the NCI-funded grants. Few investigators conduct metabolomic profiling also so that they can characterize pathways involved in the process of cancer development. The most common methods are Liquid Chromatography Mass Spectrometry (LCMS), Gas Chromatography Mass Spectrometry (GCMS) and Nuclear Magnetic Resonance (NMR). Bioinformatics tools integrating 16S data with metabolomics data are also being developed.

Table 1: Cancer types and commonly investigated viruses and bacteria with selected references.
Discussion
From the portfolio analysis of NCI-funded grants some major themes were observed. These themes trend toward the characterization of overall microbiota community structure over a disease’s time course and in relation to various exposure events. Similarly, currently research on microbiome in cancer epidemiology is being conducted to identify biomarkers of risk, progression, and prognosis of cancer. Assessing the role of select microbiota or relative composition of microbiota, in disease prevention or progression is also a current trend.
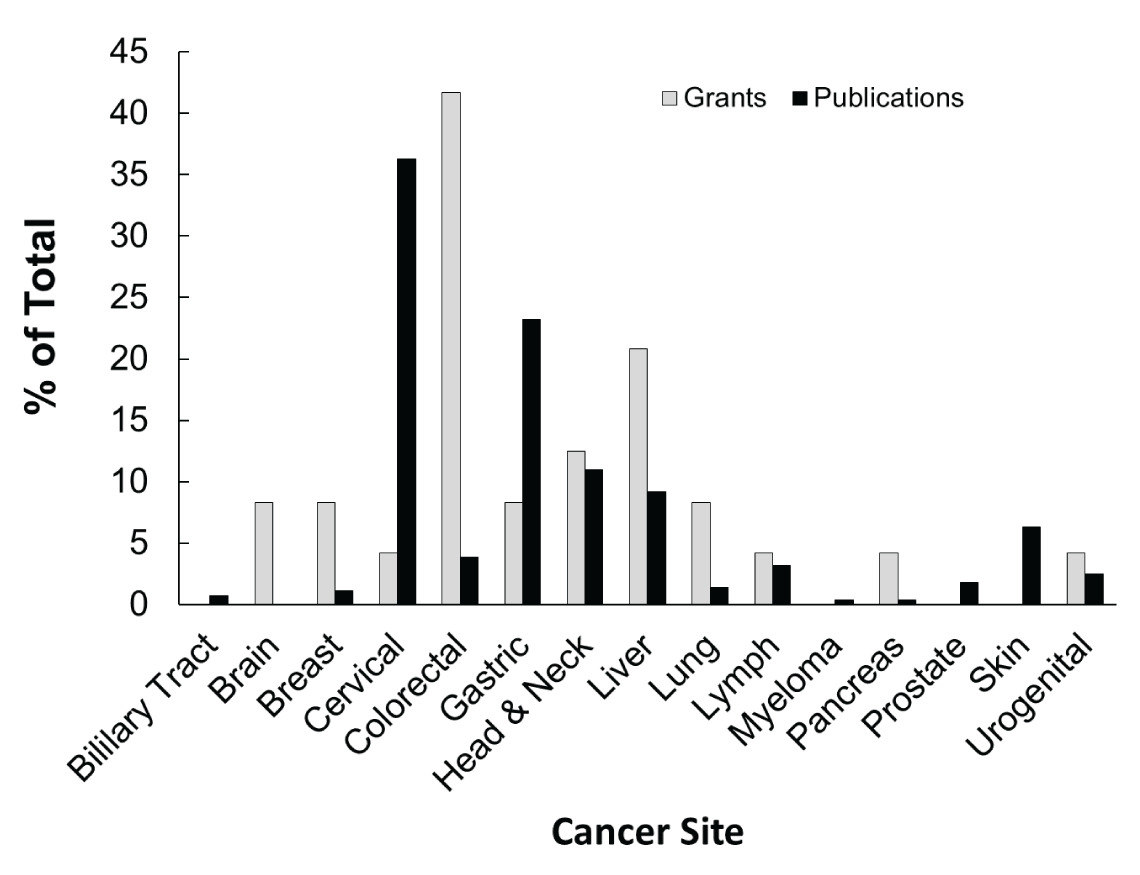
Figure 1: Percentage of the total number of NCI-supported grants or PubMed publications, categorized by cancer site, that have incorporated microbiome analysis in cancer epidemiology.
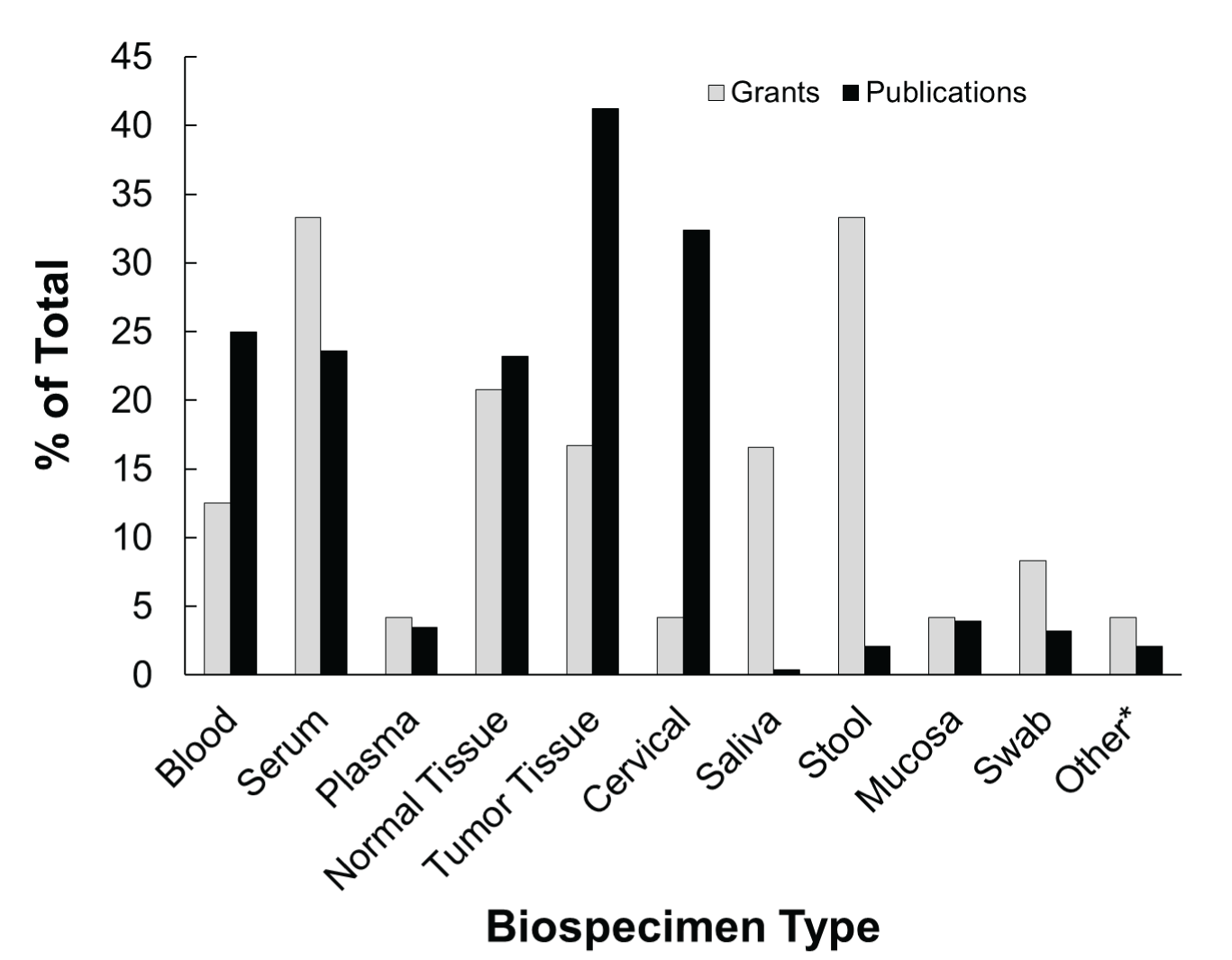
Figure 2: Most commonly collected biospecimens types for microbiome analysis in cancer epidemiology. Percentage of the total number of NCIsupported grants or PubMed publications that have utilized the different biospecimens for microbiome analysis are shown. *Other includes: Eyebrow Hair, Oral Rinse/Wash, Bile, Nasal Mucus, and Urine.
The majority of publications that were included in this analysis investigated known carcinogenic infectious agents such as HPV and cervical cancer, rather than what some might consider traditional microbiome studies. The reason for the inclusion of these studies in our analysis is in keeping with the definition of ‘microbiota’ to include viruses, which traditionally have been more commonly associated with cancer. This taken together, with the inclusion criteria of at least 100 cases, resulted in a limited number of true microbiome studies within the published literature. This demonstrates a gap in the research and also a possible starting point for inclusion of microbiome analysis in established cohorts or cohorts with existing samples. It is also recommended that cancer grants that proposed microbiome analysis and investigating novel associations in less studied cancers, such as pancreatic cancer and ovarian cancer, be keenly considered for funding.
From this analysis, some challenges and knowledge gaps have been identified and outlined in table 1. In order to successfully analyze the microbiome over time in relation to pre-cancerous states, evidence supporting the stability of various sample types over time and at various temperatures is needed in order to establish guidelines for all epidemiologic studies of the microbiome. This has been done for stool samples [20] but confirmation is needed for the various other biospecimen types [21].
Standardization of methods for reliable data integration across platforms and research centers is critical moving forwards. A large amount of sequence data already exists from microbiome research; however, this data was obtained using several different platforms and analysis softwares or pipelines available, each having inherent biases creating inaccurate data. Thus, it is critical to identify the biases for each and develop software that can correct and harmonize existing data for further research analysis. Standardization of methods, along with improvements to the reliability of available technology, will be paramount to streamlining the use of microbiome analysis in cancer epidemiology
Though much of the work with the gut microbiome and colorectal cancer has elucidated the importance of these interactions in promoting disease, a similar level of understanding for other cancers and microbiomes has yet to be achieved. In addition, the exact mechanisms occurring between the host and microbiota to influence and determine cancer progression is not yet understood for the majority of cancers as it has been for colorectal cancer. Similarly, we do not have information on potential effects resulting from interactions of the microbiota with other carcinogenic infectious agents and/or chemical agents, and how these interactions, and the resulting metabolic changes modify or determine disease progression.
Regarding the potential harm or protection against carcinogenesis that might be the effect of the microbiome, one example is that of cancerassociated infectious agents. Chronic infections caused by the hepatitis B and C viruses, human papillomaviruses (HPV), and Helicobacter pylori (H. pylori) are reported to be responsible for approximately 15% of all human cancers. Interestingly, although many of the infectious agents that have been associated with cancer such as HPV, Epstein-Barr virus (EBV), and H. pylori are highly prevalent in the world, most infected individuals do not develop cancer but remain lifelong carriers. Malignancies associated with infectious agents may result from prolonged latency as a result of chronic infections.
The microbiome is a resource that holds promise for cancer epidemiology; however, in order to reliably utilize the microbiome for all its potential, work must be done at the level of basic science research to better understand the disease promoting or protective mechanisms that are at play between the host and microbiota.
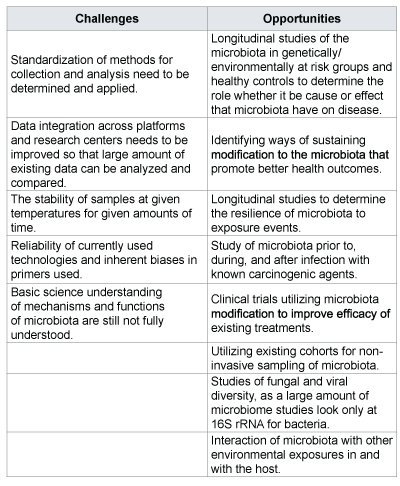
Table 2: Challenges and opportunities identified for further research.

Figure 3: Methods and technologies that are routinely being used for microbiome analysis in cancer epidemiology: The chart represents the percentage of the total number of NCI-supported grants or PubMed publications that are using the different methods and technologies. *Chromatography includes: liquid chromatography, gas chromatography, and chromatography combined with mass spectrometry. **Other includes: Giemsa Stain, histology, and clinical diagnostic tests.
Presently, it is feasible to collect noninvasive samples representative of microbiomes from various potential cancer sites over multiple time points. This practice can easily be included in epidemiologic studies, which potentially may provide information on the natural history and disease progression, identification of those at highest risk, and ultimately inform research into specific mechanisms that lead to pre-cancerous states. Studying the microbiome in this way may also provide valuable information on how variables like diet and stress contribute to cancer risk.
Opportunities for further study are presented in Table 1, which shows the commonly investigated bacteria or viruses and their associated cancer sites, may offer general starting points for investigation. Each of these cancer sites has an associated microbiome that can be sampled over time with the potential to yield information related to the natural history of a particular cancer. As a possible example of this, one might observe community shifts as a result of infection and ultimately progression to pre-cancer or cancer, allowing further risk stratification within a given population. Potential findings from longitudinal studies of the microbiomes of at risk populations may lead to the identification of better biomarkers for progression, which may allow for the implementation of more targeted and effective prevention strategies.
In conclusion, the field of microbiome analysis offers a unique opportunity for cancer epidemiology which is beginning to receive a lot attention from the scientific community. If the challenges outlined in Table 2 are addressed, the microbiome, when combined with clinical and molecular epidemiology, may identify surrogate markers of disease progression for microbe-associated cancers, and may help in better cancer diagnosis, prevention, prognosis, and treatment outcomes assessment.
We acknowledge Dr. Jessica Pierce Ph.D, Genetics and Biochemistry Branch, NIDDK, NIH for reviewing early drafts of this manuscript.
Download Provisional pdf here
All Sci Forschen Journals are Open Access