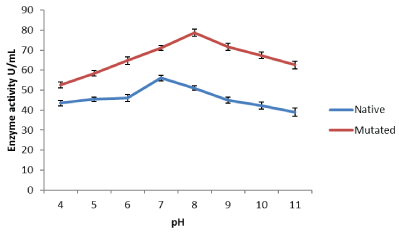
Figure 1: Influence of pH on activities of protease from native Bacillus subtilis IBL-04 and UV-90 mutant.

Mohsin Iqbal* Muhammad Asgher Fareeha Bashir
Industrial Biotechnology Lab, Department of Biochemistry, University of Agriculture, Faisalabad, Pakistan*Corresponding author: Mohsin Iqbal, Industrial Biotechnology Lab, Department of Biochemistry, University of Agriculture, Faisalabad, Pakistan, E-mail: mohsiniqbal5050@gmail.com
The present study illustrates the purification and kinetic characterization of an alkaline protease from UV-90 mutant strain of Bacillus subtilis. The enzyme was purified using four steps purification Ammonium Sulfate Precipitation, Dialysis, Ion Exchange Chromatography and Gel Filtration Chromatography. The protease produced under optimum conditions (pH 8 temperature 50°C, inoculums, 2.5 mL and fermentation time 72 hours with enzyme activity 95.89 U/mL) was 2.47-fold purified through four steps purification protocol and the purified enzyme was characterized by studying the effect of varying pH, temperature and substrate concentrations. Kinetic Studies showed that Km and Vmax values for purified protease (UV-90) were 0.03064 µM and 69.76 U/mL, respectively as compared to 0.02669 µM and 56.73 U/mL of native protease respectively. These characteristics render its potential use in biotechnological and industrial sectors.
Alkaline Protease; Bacillus subtilis; Purification; SDSPAGE; Diethyl Aminoethyl (DEAE) Ion-Exchange Chromatography; Characterization
Microorganisms are known to play a vital role in technology development for the production of intracellular and extracellular enzymes on an industrial scale. For getting maximum product yield, the selected microorganisms are grown in fermenters under optimum conditions and can be further used to make products such as enzymes, cheese, bread, biofuels and other products [1].
Most reactions in the biological system require enzymes, which act as catalysts and are essential for life. Bacillus species are the main producers of extracellular proteases, and in fermentation process on industrial scale frequently use Bacillus subtilis to produce alkaline protease. Proteases are the main enzymes produced from microbial sources, of which only few are recommended as commercial producers [2].
Bacillus subtilis is found mainly in soil and is also known as hay bacillus and grass bacillus. It is a rod-shaped organism, which can form a tough, protective endospore and can withstand extreme environmental conditions. Bacillus species are obligate aerobes or facultative anaerobe and include both free-living and pathogenic species [3].
In the previous study, the UV irradiation mutagenesis was carried out to develop a high protease yielding mutant of Bacillus subtilis IBL-04. After the selection of hyperproducing mutant strain, the physical parameters (inoculum size, incubation time, pH and temperature) were optimized by RSM (Response Surface Methodology) under CCD (Central Composite Design) to improve the production of protease. Hyperproduction of protease was obtained at optimum pH 8, temperature 50°C, inoculum size 2.5mL and fermentation time 72 hrs [4].
Later, the factors affecting culture conditions, productivity and properties of alkaline protease, it was considered of significance to purify and characterize. The enzyme was purified using four steps purification Ammonium Sulfate Precipitation, Dialysis, Ion Exchange Chromatography and Gel Filtration Chromatography. The enzyme was characterized through kinetic studies by studying the effect of varying pH, temperature and substrate concentration to explore the factors affecting their activity. In this paper, we aimed to purify alkaline protease from Bacillus subtilis and study the factors affecting the activity to present potential and possible application for industrial purposes.
The experimental and analytical work was carried out in industrial biotechnology laboratory, department of biochemistry, University of Agriculture, Faisalabad. All chemicals used in this study were of analytical grade. These chemicals were purchased from Sigma-USA and Oxide, UK through local agents.
The bacterial strain Bacillus subtilis was obtained from Industrial Biotechnology lab (IBL), Department of Biochemistry, University of Agriculture, and Faisalabad. Bacillus subtilis was grown on nutrient agar slants at 37°C for 24 h and then preserved at 4°C for 15 days [5].
Protease production was carried out by [6]. It was further enhanced by optimizing the culture conditions by RSM (Response Surface Methodology) under CCD (Central Composite Design). Hyperproduction of protease was obtained at optimum pH 8, temperature 50°C, inoculum size 2.5mL and fermentation time 72 hrs [4].
Protease enzyme activity was measured by the addition of casein in phosphate buffer (pH 7) at 37°C. The oxidation of casein was determined at 660 nm using a spectrophotometer. Enzyme activities were expressed in U/mL [7,4].
Formula:
\[Units/mL\,enzyme = \frac{{\mu mol\,tyrosine\,equivalents\,released \times total\,volume\,of\,assay}}{{A \times B \times C}}\]
A = volume of enzyme used
B = Time required for assay (20min)
C = Volume used in colorimetric determination (1ml)
Crude enzyme was subjected to ammonium sulfate precipitation technique. Crude enzyme extract obtained from a culture of BS-90 mutant was centrifuged at 8000 rpm for 10 min. The cell-free supernatant/filtrate was brought to 80% saturation. Ammonium sulfate crystals were gradually added in order to achieve 80% saturation. It was again kept for overnight at 4°C and centrifuged 8000 rpm for 10 min. After centrifugation, the supernatant was discarded. Sediments were dissolved in a minimal volume of 50 mM phosphate buffer of pH 7 [8]. After ammonium sulfate treatment, the solution was dialyzed against assay buffer several times to salt out ammonium sulfate completely. Protease enzyme activity was measured by spectrophotometer before and after dialysis along with protein content [8].
Ion exchange chromatography: After dialysis, the protease was further purified by Ion-exchange chromatography which ultimately separates proteins based on molecular charge. The column of DEAE-cellulose (Diethyl aminoethyl cellulose) was prepared by adding DEAE-cellulose resin in distilled water and heating in a water bath for 5 hours at 95°C with continuous stirring. After that arrange the column vertically and transferred the prepared slurry into the column, and let the column undisturbed overnight. Then washed the column with 30 mL of 0.5N NaOH solution followed by washing with 30 mL of 0.5N solution of HCl.1 mL solution was then poured into the column with 50 mM phosphate buffer (pH 7). After washing the column was equilibrated with phosphate buffer with a pH of 7 and the adsorbed protein was eluted with a stepwise NaCl gradient prepared in 50 mM phosphate buffer (pH 7). 1 ml fractions were collected wherein eluted fractions were monitored at 280 nm. The peak with the highest protease activity was dialyzed against 50 mM phosphate buffer (pH 7).Protein concentration and protease activity was determined by routine assay methods [9].
Gel filtration chromatography: Gel filtration technology was used for the detection of discrepancy retention of molecules (inversely related to size) due to entry into pores of the gel. A column of Sephadex G-100 was prepared by adopting the method of [8]. Two mL protease sample after ion exchange chromatography was loaded on Sephadex G-100 column to get further purification to homogeneity level. 50 mM phosphate buffer of pH 7 and elution of fractions of protease was collected and monitored the absorbance at 660 nm by a spectrophotometer.
The purified enzyme was characterized through a kinetic study by studying the effect of pH, temperature, substrate concentration along with the determination of km and Vmax values using Line Weaver-Burk plot [8].
Effect of pH: In order to determine the pH optima for protease, the reaction mixture was incubated for 15 min in buffers of pH 4.0-11.0. After incubation, the enzyme assay was performed using standard assay protocol. The buffers used were: acetate-buffer, pH 4.0, 5.0; phosphate-buffer, pH 6.0, 7.0 and 8 and glycine-sodium hydroxide buffer, pH 9.0, 10.0 and 11.0.
Effect of temperature: The protease was incubated at different temperatures i.e. 30°C -85°C for one hour at optimum pH before running the routine protease assay to determine the optimum temperature for protease activity.
Effect of varying substrate concentration: Effect of substrate concentration on activities of protease was studied at optimum pH and temperature using varying concentration of casein in mM. Linewear’ s-Burk plot (reciprocal) were constructed between 1/S and 1/Vo and kinetic parameters of MichaelisMenten (KM and Vmax) were determined [10].
For ammonium sulfate precipitation, the salt was added to the crude extract to 80% saturation at 4°C and 1.32 folds purification was achieved. Protease was 1.64 times purified by dialysis. In the column prepared with the dimethyl aminoethyl cellulose resin (anion exchange column), sample solution was loaded. The column was calibrated with 20-30 mL phosphate buffer (pH 7) and 2mL crude protease enzyme was loaded. Subsequently, gel chromatography column was packed by means of Sephadex-100. Protease activity fractions isolated by DEAE have purification fold of 1.84 whereby Sephadex column chromatography 2.47 times higher purification was attained (Table 1).
| S. No | Purification Steps | Volume of Sample (mL) | Total Enzyme Activity (IU) | Total Protein Content (mg) | Specific Activity (U/ mg) | Purification Fold |
| 1 | Crude Enzyme | 300 | 95.89 | 18.21 | 5.26 | 1 |
| 2 | (NH4)2 SO4 Precipitation | 30 | 93.76 | 13.43 | 6.98 | 1.32 |
| 3 | Dialyzed Enzyme | 29 | 87.23 | 10.1 | 8.63 | 1.64 |
| 4 | DEAE-Cellulose | 28 | 82.89 | 8.56 | 9.68 | 1.84 |
| 5 | Sephadex G-100 | 15 | 76.73 | 5.89 | 13.02 | 2.47 |
Table 1: Purification scheme for the Protease produced by mutant strain UV-90 of Bacillus subtilis IBL-04.
The substantial loss on the whole yield could be correlated with the low protease stability in the highly acidic medium. Thus, in order to enhance the yield, future prospective might examine chromatographic techniques at its appropriate pH values. Therefore, it is essential to accentuate that the yield gained for the purification procedures in diver’s reports, which use different strategies is extremely inconsistent and it associated to the specific properties for each crude and purified enzyme [9].
Effect of pH on protease enzyme: Protease enzyme was incubated at varying pH ranging from 4-11 before using. The optimal pH for the mutant protease enzyme was 8.0 whereas the enzymes were less reactive at low pH and were more reactive in alkaline pH. It had been noticed that enzyme activity can be enhanced by changing the confirmation of enzyme after entrapment [11].
The alkaline protease from UV-90 mutant was completely stable in a large alkaline pH range (4-11) and presented an optimum activity for 78.73 U/mL at a pH value of 7 whereas any further increase in pH up to 11 showed decreasing trend in activity (Figure 1). The pH optima for the alkaline protease of Bacillus species have been reported to vary from 4-11. Our results are in close agreement with [12] who reported that protease enzyme was active in the range of pH 7-13 with an optimum pH 10 and presented an optimum activity 126 U/mL.

Figure 1: Influence of pH on activities of protease from native Bacillus subtilis IBL-04 and UV-90 mutant.
In view of other investigators, Johnvesly et al, (2002) [13] reported that, the optimal protease activity produced by Thermoalkaliphilic Bacillus sp. JB-99 was observed at pH 11. So, this enzyme showed stable activity under alkaline conditions. Similarly, the properties of protease from Bacillus sphaericus strain C3-41 showed optimum activity at pH 11.0. The enzyme was stable up to pH5.0-12.0 [14].
Effect of temperature on protease enzyme: The experiment was conducted to determine the effect of different incubation temperatures (30-85°C) on the purified protease enzyme. It was observed that mutant protease enzyme was stable up to 70°C. Whereas highest protease activity from native strain was obtained at 45°C. It was concluded from the experiment that at temperatures higher than 70°C enzyme starts to losses its activity rapidly (Figure 2). For a variety of industrial applications relatively high thermostability is an attractive and desirable characteristic of an enzyme.
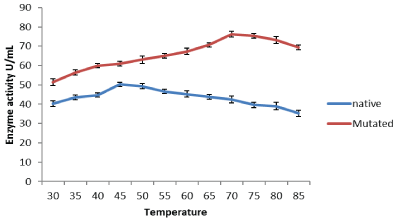
Figure 2: Effect of varying temperature on activities of protease synthesized from native and mutant strain of Bacillus subtilis IBL-04.
Haddar et al. [15] reported that the constant and highest protease enzyme activity was observed at a temperature between 25-60°C when using casein as substrate. The optimum temperature for purified native alkaline protease was observed at 45°C. Most reported protease enzymes presented their maximum rates in the range of 30-85°C [12]. Residual protease activity was determined at different temperatures and was maximal at 40-50°C, although considerable residual activity (50%) was observed even after incubation at 80°C for 30 min [7]. Similarly, Johnvesly et al. [13], reported that the optimum temperature for protease activity was 70ºC produced by thermoalkaliphilic Bacillus sp. JB-99.
Effect of substrate concentration on protease enzyme: The Km and Vmax for native and mutant purified protease were determined by varying the concentration of (casein) substrate. The enzyme activities were measured under standard assay conditions and results were used to build the reciprocal plot.
The reciprocal graph of protease activity (1/V) was plotted against reciprocal of each substrate concentration (Figure 3). Kinetic Studies showed that Km and Vmax values for mutant purified protease were 0.03064 µM and 69.76 U/mL, respectively, when using casein as substrate. Whereas the Km and Vmax values for native protease were 0.02969 µM and 56.73 U/mL. The relationship between the rate of reaction and concentration of substrate depends on the affinity of the enzyme for its substrate expressed as Km of protease enzyme [16,17].
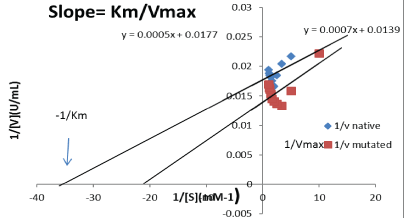
Figure 3: Line weaver Burk Reciprocal plot of Protease enzyme for determination of Km and Vmax values for native and UV-90 mutant of Bacillus subtilis.
Ahmed et al. [12] reported that the catalytic properties, Km and Vmax values of purified alkaline protease from Bacillus subtilis were 58 μM and 148 U/mL, respectively. An enzyme with low Km has a greater affinity for its substrate. Alkaline protease is highly substrate specific and exhibit maximum activity towards casein as substrate. It was reported that protease has a high level of hydrolytic activity against casein as substrate and poor to moderate hydrolysis of BSA and egg albumin, respectively [18]. According to Patel et al. [19] results, the Km and Vmax of protease were 0.153 g/100mL and 454 U/mL, respectively.
From the present study, it is concluded that the identified species, Bacillus subtilis possesses good protease activity. The study has also standardized the growth parameters of bacteria for the maximum enzyme production, which can be effectively used in the large-scale production of protease for commercial purposes. The optimum temperature and pH for UV-mutant protease were determined at 50°C and 8.0 respectively. The protease was purified by ammonium sulfate precipitation, Dialysis, Ion Exchange Chromatography and Gel Filtration Chromatography. The thermoalkalophilic protease will be used for the various purposes in detergent industries, food industries and pharmaceutical industries owing to stability at high temperature and pH along with high Vmax and substrate specificity.
Download Provisional PDF Here
Article Type: RESEARCH ARTICLE
Citation: Iqbal M, Asgher M, Bashir F (2018) Purification and Kinetic Characterization of Alkaline Protease for UV-90 Mutant of Bacillus Subtilis. J Biochem Analyt Stud 3(1): dx.doi.org/10.16966/2576-5833.112
Copyright: © 2018 Iqbal M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Publication history:
All Sci Forschen Journals are Open Access