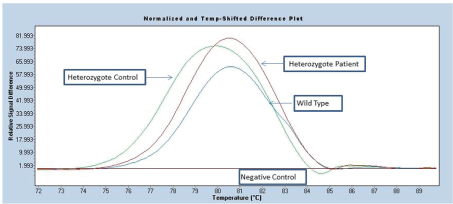
Figure 1: HRM assay for FLT-ITD mutation


Dilara Fatma Akin1* Mine Mumcuoglu1 Deniz Aslar Oner1 Mehmet Ozturk2 Ustun Ezer2 A Emin Kurekci2 Nejat Akar3
1Cancer Research and Genetics Laboratory, Losante Children’s and Adult Hospital, Ankara, Turkey*Corresponding author: Dilara Fatma Akin, Cancer Research and Genetics Laboratory, Losante Children’s and Adult Hospital, Ankara, Turkey, Tel: +905363026816; E-mail: dilarafatmaakin@gmail.com
Objective: Important role of FMS-related tyrosine kinase (FLT3) in development of hematopoietic and immune system has been known. Studies have shown that FLT3 mutations play critical role at the pathogenesis acute myeloid leukemia (AML). FLT3- internal tandem duplications ((FLT3-ITD)) reported as a significant prognostic marker for pediatric AML. But it is unclear for acute lymphoblastic leukemia (ALL), very few data are presently available in ALL. Studies investigating FLT3 mutations in Turkish pediatric acute leukemia are very few. Also FLT3-D835 mutation status in pediatric leukemia has not been explored very well. Therefore, our aim in this study was to screen and analyze FLT3 mutations in Turkish pediatric acute leukemia patients.
Materials and Methods: Study population was consisted of 27 pediatric patients for (FLT3-ITD) mutation and of 183 pediatric patients for FLT3-D835 mutation who were diagnosed with acute leukemia. (FLT3-ITD) mutation screening performed by using fluorescence HRM (High Resolution Melting) analysis in real-time Polymerase Chain Reaction (PCR) method. FLT3-D835 variation was detected with Restriction Fragment Length Polymorphism (RFLP) method.
Results: We detected (FLT3-ITD) mutation in 33.3% of our study group (n=27). FLT3-D835 screening showed 7.6% of patients were carried heterozygote genotype. Statistical analysis showed that FLT3-D835 heterozygote mutation was correlated with diagnosis and risk groups. But same correlation was not identified for (FLT3-ITD) mutated samples. Also we found no correlation between FLT3 mutations and clinical characteristics of patients.
Conclusion: We observed high percentage of FLT3 mutations in Turkish pediatric acute leukemia patients compare to previous studies. Our results show the importance of FLT3 mutations in pediatric acute leukemia patients as an important prognostic and diagnostic marker.
Childhood acute leukemia; (FLT3-ITD); FLT3-D835; Tyrosine kinase
AML: Acute Myeloid Leukemia; ALL: Acute Lymphoblastic Leukemia; HRM: High Resolution Melting; PCR: Polymerase Chain Reaction; RFLP: Restriction Fragment Length Polymorphism
The FMS-related tyrosine kinase (FLT3) is a receptor tyrosine kinase which has an important role proliferation, survival and differentiation of hematopoietic progenitor cells [1,2]. FLT3 is one of the class III tyrosine kinase receptors which consist of five immunoglobulin-like domains in extracellular region, an intracellular juxtamembrane (JM) domain, two kinase domains with kinase insert and a C-terminal tail [3]. FLT3 gene is located at chromosome 13 (13q12) and has 24 exons [4]. High expression of FLT3 has been shown in acute myeloid leukemia (AML), acute lymphocytic leukemia (ALL) and in the blast crisis of chronic myeloid leukemia (CML) [5]. Rosnet et al. [6] has also shown that FLT3 is expressed on leukemic blast in AML and B-ALL cases. Two types of FLT3 mutation have been described in leukemia: FLT3 internal tandem duplications ((FLT3-ITD)) and tyrosine kinase domain (TKD) mutation. A tandemly duplicated sequence was located in JM domain of FLT3 coded by the exons 14 and 15. TKD mutation of FLT3 comprises mutations in the second tyrosine kinase domain especially at codon D835. All these mutations generate changes in the amino acid sequences and cause constitutive activation of FLT3 kinase activity [7]. (FLT3-ITD) mutation prevalence for pediatric AML has been described in 11-33% and identified as a significant independent prognostic factor for poor outcome [8-12]. (FLT3-ITD) allelic ratio (mutant-wild type ratio is greater than 0.4) was also found important prognostic factor for relapse in pediatric AML [13]. It has been suggested that (FLT3-ITD) detection should be considered and performed as a routine test at the diagnosis of AML and management of the therapy [9]. Other type of FLT3 mutations has been seen at a lower frequency compare to (FLT3-ITD) mutations in acute leukemia patients. There are very few studies which investigate FLT3 mutations in Turkish pediatric leukemia patients [14,15]. Therefore in our study, we aimed to screen (FLT3-ITD) and TKD mutations by HRM and PCR analysis and finding possible genetic markers for molecular leukemia.
Study population was consisted of 27 patients for (FLT3-ITD) mutation and of 183 patients for FLT3-D835 mutation their age’s between 1 and 15 years who were admitted to Losante Pediatric Hematology-Oncology Hospital, Ankara, Turkey with the diagnosis of acute leukemia. Blood samples were taken to the EDTA-containing tubes and DNA was extracted from peripheral blood leukocytes with MagNA Pure automatic DNA isolation instrument (Roche Diagnostics, Manheim, Germany). Informed consents were collected from the patient’s parents prior to the study.
For (FLT3-ITD) mutation: (FLT3-ITD) variation was screened with real time PCR (Genes-4u FLT3-ToolsetTM Roche Diagnostics, Gmbh, Manheim, Germany) using fluorescence HRM analysis based on genotype profiles. Different plots were created by selecting negative controls as the base-line. Therefore, fluorescence of the all other samples was diagramed relative to this sample. Fluorescence signals were analyzed and significant differences used as indicators of mutations [16].
For FLT3-D835 mutation: Amplification of gene was performed by PCR. Primers used in FLT3-D835 mutation were as follows: forward primer: 5’-CCG CCA GGA ACG TGC TTG-3’; reverse primer: 5’-GCA GCC TCA CAT TGC CCC -3’. Primers used in FLT3-D835 mutation were designed as previously described by Moreno et al. [17]. At codon 835 an aspartate amino acid is encoded, providing a recognition site for restriction enzyme EcoRV; as such, mutants can be detected via the loss of this enzyme restriction site.
Then amplified fragments were digested with appropriate restriction endonucleases (EcoRV, Fermentas, Lithuania) and sequencing of different band profiles was performed by sequencer (Beckman-Coulter CEQ 2000 XL DNA Analysis System, USA).
Statistical analyses were performed by using The Statistical Package for Social Sciences (SPSS) version 20 software. Correlations between FLT3 mutations and either diagnosis or risk groups were analyzed by using One way ANOVA test. Mann-Whitney test was applied to determine correlations between FLT3 mutations and clinical and laboratory characteristics of patients. P values less than 0.05 were referred as statistically significant.
Twenty seven children diagnosed with acute leukemia were included to screening study of (FLT3-ITD) mutation. Screening performed by using fluorescence HRM analysis (Figure 1). Figure 1 displays evaluation of HRM analysis for (FLT3-ITD) mutations.

Figure 1: HRM assay for FLT-ITD mutation
Detailed clinical laboratory data of 27 children is given at table 1. We determined (FLT3-ITD) mutation in 9 (33.3%) of them. One (3.7%) of the 27 patients carried both (FLT3-ITD) mutation and FLT3-D835 heterozygote phenotype. Distribution of (FLT3-ITD) positive patients according to their diagnosis were as follows: AML (n=5); B-ALL (n=3); Pre B-ALL (n=1). Median age for ITD positive patients was 8 (range: 2-13) and for ITD negative patients was 5.25 (range: 1.2-10). Percentage of male patients was higher than female patients in both (FLT3-ITD) positive (male: 66.6%; female: 33.3%) and (FLT3-ITD) negative (male: 61.1%; female: 38.8%) samples. Median white blood cell count (WBC) in (FLT3-ITD) positive patients was 12600 × 109 L-1 and in (FLT3-ITD) negative patients was 12150 × 109 L-1. Median Hemoglobin (Hb) level in (FLT3-ITD) positive patients was 9.3 g dL-1 and in (FLT3-ITD) negative patients was 8.3 g dL-1. Median platelet (PLT) in (FLT3-ITD) positive patients was 75000 × 109 L-1 and in (FLT3-ITD) negative patients was 26000 × 109 L-1. Median bone marrow blast in (FLT3-ITD) positive patients was 98% and in (FLT3-ITD) negative patients was 100%. Median CD34 in (FLT3-ITD) positive patients was 10% and in (FLT3-ITD) negative patients was 35%. Diagnostic distribution and summary of clinical features of patients are shown at table 2.
Characteristics |
(FLT3-ITD)Positive patients |
(FLT3-ITD)Negative patients |
No patients |
9 |
18 |
Diagnosis, no. (%) |
||
AML |
5 (55.5) |
4 (22.2) |
Pre B-ALL |
1 (11.1) |
8 (44.4) |
B-ALL |
3 (33.3) |
2 (11.1) |
T-ALL |
0 |
2 (11.1) |
JMML |
0 |
1 (5.5) |
ALL |
0 |
1 (5.5) |
Median age |
8 (2-13) |
5.25 (1.2-10) |
Sex, no. (%) |
||
Male |
6 (66.6) |
11 (61.1) |
Female |
3 (33.3) |
7 (38.8) |
Median WBC count ( × 109 L-1) |
12600 |
12150 |
Median Hb (g dL-1) |
9.3 |
8.3 |
Median PLT ( × 109 L-1) |
75000 |
26000 |
Median BM Blast (%) |
98 |
100 |
Median CD34 (%) |
10 |
35 |
T(9;22) |
||
Positive |
1 |
3 |
Negative |
8 |
15 |
Table 1: Clinical characteristic and mutational status of 27 patients included FLT3 mutation screening
Table 2: Clinical characteristic and mutational status of 27 patients included FLT3 mutation screening WBC: White blood cell; Hb: Hemoglobin; PLT: platelet; BM: Bone Morrow; WT: Wild Type; HR: High Risk; ST: Standard Risk; MR: Moderate Risk, NA: Not Available.
Screening for FLT3-D835 mutation was performed on 183 children who were diagnosed with either acute lymphoblastic or myeloid leukemia. RFLP method was applied for this screening (Figure 2). Experiment results showed that 14 of 183 children (7.6%) were carried heterozygote genotype. Sequencing of the gene of heterozygous patients revealed a heterozygous “C/A” variation at c.32705. Nine of 14 heterozygote patients (64.2%) were diagnosed with Pre B-ALL. Two of 14 heterozygote patients (14.2%) were diagnosed with B-ALL. Distribution based on their diagnosis of the remaining heterozygote patients (three out of 14) were T-ALL, ALL and JMML.
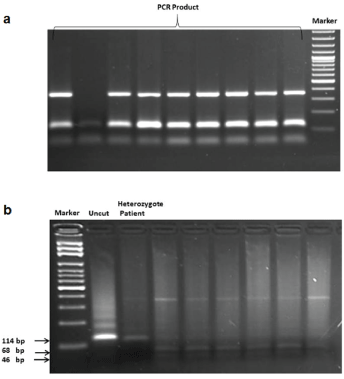
Figure 2: a) PCR amplification of FLT3-D835 region. b) Digestion products of amplified fragments for detection of FLT3-D835 mutation.
Experiment results showed significant correlation between FLT3-D835 heterozygote mutation and diagnosis (p=0.003) and also risk groups (p=0.045). There was no correlation between (FLT3-ITD) mutation and either diagnosis or risk groups. We explored the possibility of correlation between FLT3 mutations and clinical characteristics of patients (age, WBC, PLT, Hb, CD34 and BM blast) and we found no correlation.
Significance of FLT3 in normal lymphohematopoietic stem/progenitor cell function has been known. Hence FLT3 mutations play critical role in pathogenesis of leukemia. (FLT3-ITD) mutation is one of the most common mutations in adult AML, occurring approximately 20-27% of patients [9,18-21]. In pediatric AML patients, frequency of (FLT3-ITD) mutation is lower (5.3-22.2%) than adult patients [20]. There is no too many studies that investigate FLT3-D835 mutations in pediatric acute leukemia patients. Liang et al. [22] investigated FLT3-D835 mutation in pediatric AML patients and found 3.3% frequency of this mutation. Nasiri et al. [23] studied FLT3-D835 mutation in 100 pediatric acute leukemia patients and found frequency of this mutation as 3.7% in AML and 1.3% in ALL patients. Studies performed in Saudi pediatric ALL patients and Serbian pediatric AML patients showed frequency of FLT3-D835 mutation as 2.1% and 9.5%, respectively [24,25]. To the best of our knowledge, there are only two studies that were investigated FLT3 mutations in Turkish pediatric acute leukemia patients [14,15]. Ozbek et al. [14] reported FLT3- ITD mutation rate as 4% and they observed FLT3-D835 point mutation heterozygosity in only 1 patient (2.0%). Karabacak et al. [15] found FLT3- ITD mutation in 7.5% in ALL patients and 22.5% in AML patients. They didn’t identify FLT3/TKD mutation in any of their patients. In our study, we found (FLT3-ITD) mutation in 55.5% of AML diagnosed patients and in 22.2% of ALL diagnosed patients. FLT3-D835 heterozygosity was observed 7.6% of our study group. Therefore, our results showed higher (FLT3-ITD) and FLT3-D835 mutation frequencies than Ozbek et al. [14] and Karabacak et al. [15] Beside Ozbek et al. [14] used PCR method to screen (FLT3-ITD) mutation. But we prefer to use HRM analysis to identify (FLT3-ITD) mutation. HRM analysis is more sensitive method compare to PCR [26]. Our high (FLT3-ITD) mutation rate was possibly due to our sensitive analysis method. Additionally our results show differences and significance compare to Ozbek and Karabacak studies in terms of study group composed of not only AML patients and also ALL patients and we observed 7.6% FLT3-D835 heterozygosity in our leukemia group.
One patient in our study group showed (FLT3-ITD) mutation and FLT3-D835 point mutation heterozygosity together. This patient is eight year old girl diagnosed with B-ALL and classified as a high risk group. She has 12000 ( × 109 L-1) WBC count, 10.5 (g dL-1) Hb, 115000 ( × 109 L-1) PLT count, 18% BM blast and 8% CD34. Cytogenetic tests showed trisomy 8 in her bone morrow samples. She entered first remission at 28th day of her chemotherapy treatment. But 18 months later she diagnosed with relapse. Reported double mutation cases have been very few [14,17,27]. These studies performed on AML patients. Double mutation carrying patients died in reported studies either during treatment or after relapse [17]. To the best of our knowledge, this is the first case with double FLT3 mutation, trisomy 8 and ALL. Beside FLT3 mutations, trisomy 8 may play role of this patient’s poor prognosis [28]. Therefore double FLT3 mutations in ALL could be associated with bad prognosis and relapse like AML patients.
In our study, we found significant correlation between FLT3-D835 heterozygote mutation and diagnosis (p=0.003) and also risk groups (p=0.045). But similar correlation was not detected between (FLT3-ITD) mutation and either diagnosis or risk groups.
Ozbek et al. [14] reported no correlation between FLT3 gene mutations and age, gender, WBC count, blast cell rate or FAB classification in their study group composed of AML patients. Karabacak et al. [15] concluded that there is no significant relationship between laboratory results and FLT3/ITD positivity in both ALL and AML patients. But they found association between FLT3/ITD positivity and increase in age. We found no correlation between FLT3 mutations and clinical characteristics of patients (age, WBC, PLT, Hb, CD34 and BM blast) in our study similar with Özbek’s and Karabacak’s study. But some studies found positive association with high WBC count and high percentage of bone morrow blast cells in (FLT3-ITD) positive samples [9,23].
Our statistical analysis showed significant correlation between FLT3-D835 heterozygote mutation and diagnosis (p=0.003) and also risk groups (p=0.045). This result showed that FLT3-D835 mutation can be used as a prognostic marker in ALL and helpful information for the diagnosis. It is known that (FLT3-ITD) mutation is a poor prognostic marker for acute leukemia. But we didn’t find similar correlation between (FLT3-ITD) mutation and either diagnosis or risk groups. This result could be due to our small and mixed study group.
Cytogenetic prognostic factors are preferred more in clinical use. But there is a need to enhance analysis and clinical use of molecular factors such as mutational status FLT3, MLL, NPM1, CEBPA especially in cases with normal karyotype.
Our study is one of the few studies that analyze FLT3 mutational status in Turkish pediatric acute leukemia patients. It also points out the importance of FLT3-D835 mutations in ALL prognosis.
The authors declare that they have no conflict of interest, including specific financial interests, relationships, and/or affiliations relevant to the subject matter or materials included.
Download Provisional PDF Here
Article Type: Research Article
Citation: Akin DF, Mumcuoglu M, Oner DA, Ozturk M, Ezer U, et al. (2016) Screening of FLT3 Gene Mutations (Flt3-Itd and D835) in Turkish Childhood Acute Leukemia Patients. J Mol Med Clin Appl 1(1): doi http://dx.doi.org/10.16966/2575-0305.104
Copyright: © 2016 Akin DF, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Publication history:
All Sci Forschen Journals are Open Access