Introduction
The Ranaviruses (family: Iridoviridae) affect lower vertebrates, including amphibians [1,2]. Some Ranavirus species/strains infect most, if not all, potential amphibian hosts in a community [3-6]. However, not all strains of Ranavirus appear to exploit a broad range of host species [7] and some may even co-evolve with single host species (e.g. Ambystoma tigrinum virus and Ambystoma tigrinum) [8].
Ranavirosis emerged in the southeast of England, United Kingdom, in populations of common frogs (Rana temporaria) in the mid- to late-1980s [9]. Since then, infection with Ranavirus has been described in other UK amphibians [common toads (Bufo bufo)] [10]; invasive common midwife toads (Alytes obstetricans) and common newts (Lissotriton vulgaris), [11] which in some cases are comparably resistant to the development of disease [7, 12]. Even in common frogs that develop ranavirosis, there is some evidence that these diseases do not always result in death; for example, healed sores, consistent with ranavirosis, have been observed in wild populations of common frogs [12-14]. The interpretation of these patterns would be improved by determining whether the Ranavirus variants affecting common frogs are related to those that affect other English amphibians. This information would form the basis for the analysis of transmission routes and co-evolution of virus and host, to understand host range and how infection dynamics in English amphibian communities is regulated [15]. Early investigations into the relationship of Ranavirus isolates from common frogs and common toads based on a partial sequence of the major capsid protein (MCP) gene revealed limited, and presumably non-functional genetic differentiation [10]. This is not surprising as the MCP region is highly conserved and has previously shown to be relatively uninformative for phylogenetic reconstructions of closely related viruses [16,17]. Evidence for local adaptation of Ambystoma tigrinum (ATV)-like Ranaviruses in the western USA [16] was only detected using other, more phylogenetically informative regions. Many studies are now beginning to use a multi-gene or expanded sequence approach to Ranavirus phylogenetics [15,18].
In this study we investigated the phylogenetic relationships between 24 Ranavirus isolates from three different English amphibian species, two of which may be alternate (or reservoir) hosts of the pathogen. Firstly, we used BLAST to examine the homologies with other Ranaviruses. Then, we created phylogenetic trees using two approaches: firstly, one tree constructed from concatenated nucleotide sequences of two partial genes and, secondly, a separate tree was made for each sequence, based on the predicted protein sequences. We then explore the relationships supported by the trees and infer their possible meaning in the context of other Ranavirus research.
Materials and Methods
For isolating and culturing virus we used fat head minnow (FMH; Pimephalespromelus) cells from the European Collection of Cell Cultures (No. 88102401, ECACC, Oxford, UK). The cells were propagated at 25°C in Eagle’s Minimum Essential Media (EMEM, Sigma–Aldrich, Andover, UK), supplemented with 1% L–glutamine (Sigma–Aldrich, Andover, UK), 0.005% Penicillin–Streptomycin (Sigma– Aldrich, Andover, UK), 0.005% Nystatin (Gibco, Invitrogen, Paislely, UK), and 10% Research Grade Fetal Bovine Serum (Hyclone, Perbio Science, Northumberland, UK).
Ranavirus-positive tissue samples were obtained from individuals that had been examined by Duffus et al. [11,19]. To isolated the virus, a small piece of hepatic tissue was homogenized in 15-20 mL isolation media (0.01% FBS, 0.01% Penicillin–Streptomycin, 0.005% Nystatin, 5X 10-4% Gentamycin and 0.01% L-glutamine) using an ultra-Turrax tube drive (IKA-Werke GMBH & Co. KG, Staufen, Germany). The suspension was then filtered using a sterile 0.22 µL syringe filter and syringe or a 50 mL Steriflip© unit with a 0.22 µL filter (Millipore, Hertfordshire, England). The filtered homogenate was then added to one 75 cm2 or split between two 25 cm2 flasks containing a confluent layer of FMH cells. The flasks were monitored daily for the development of viral plaques. When cells had detached from 90-95% of the flask, the virus suspension was harvested and filtered with a sterile 0.22 µL syringe filter and syringe before being aliquoted into 1.5-2 mL cryovials and frozen at -80°C.
Viral isolates were passaged again using confluent 75 cm2 flasks of FMH cells. This second passage of each viral isolate was used to ensure that all isolates had experienced comparable culture conditions. Each flask was inoculated with 100 µL to 1 mL of the viral isolate and contained 25 mL of maintenance media (EMEM, supplemented with 1% L–glutamine, 0.005% Penicillin–Streptomycin, 0.005% Nystatin, and 1% Research Grade Fetal Bovine Serum).
Flasks were maintained at 25°C in a cooling incubator and monitored daily for plaque formation. The virus-cell suspension was harvested when no cells were left adhering to the bottom of the flask.
DNA from the virus suspension was extracted using DNEasy Blood and Tissue Extraction Kits (QIAGEN, Crawley, West Sussex, UK). The protocol was modified in the following manner: we used 300 µL of virus and cell suspension, the virus suspension was not spun down and PBS was not used. The extracts were screened for the presence of viral DNA following the methods described by Duffus et al. [19], with the exception that 50 µL PCR reactions were used: 25 µL Multiplex Mix, 5.3 µL of each forward and reverse primer, 12.4 µL of distilled water, and 2.0 µL of template DNA. For ORF 57r, 25 µL reactions were used for sequencing. The primers used for the MCP detection and analysis were from [3] and those for ORF 57r were from [16]. The MCP was chosen because it is commonly used by many authors to determine the phylogenic relationships of Iridoviruses on large and small scales, despite the fact that it may not be particularly phylogenetically informative [10,20,21]. The locus denoted as open reading frame (ORF) 57r was chosen because of the availability of comparison sequences and because of its previous use [16], in combination with other ORFs, to examine local adaptation in ATV-like viruses in the western USA. The PCR products were cleaned up using a polyethylene glycol (PEG) precipitation (protocol from the Santos Lab, Auburn University, USA). A portion of the PEG precipitation products were run out on a 1.5% agarose gel (stained with ethidium bromide) to ensure the presence of target DNA. Sequencing was performed by COGENICS (now Beckman Coulter Genomics UK).
BLAST analysis
All nucleotide sequences were analyzed using BLAST searches to identify which published Ranavirus sequences they were most similar to. A second set of BLAST searches were performed to determine the similarity of each of the partial sequences to FV3 (Accession Number AY58484.1), the type species of the genus Ranavirus.
Phylogenetic analysis
Partial MCP sequences were aligned in MEGA6 [22] using Clustal W [23]. Initial sequences of approximately 500 base pairs were trimmed to 283 base pairs after alignment due to the short sequence obtained from the BUK 3 isolate. The MCP from an ATV isolate from Utah (Accession Number AY548312.1) was used as the outgroup. ORF 57r sequences were aligned and trimmed to partial sequences of 419 base pairs. The partial ORF 57r gene from an ATV isolate from Utah (Accession Number EU512332) was selected as the outgroup. The trimmed MCP and ORF57r sequences from the UK as well as the ATV isolates from the USA were joined end to end and aligned using MAFTT [24]. In MEGA 6, a nucleotide substitution model optimization was run and it was determined that the Kimura 2 parameter model was the best fit. A maximum likelihood (ML) tree was build using the best fit model for the concatenated DNA sequences and Bootstrap analyses with 1000 replicates were performed. The trees were condensed so that branches with less than 50% support were eliminated.
Phylogenetic trees were also built for both MCP and ORR57r partial DNA sequences. Sequences were aligned in MAFTT, imported into MEGA 6 and nucleotide substitution model optimizations were run for each set of partial sequences. Maximum likelihood trees were built using 1000 Bootstrap replicates and branches with less than 50% support were condensed. ATV isolates (see above for Accession Numbers) were again used as the outgroups.
In addition to the DNA sequence phylogenetic analysis, trees were built using the predicted protein sequences of both partial MCP and ORF 57r sequences. The partial MCP sequence protein data was analyzed using the JTT model of amino acid substation [25] that was found to be the best fit for our data using MEGA 6. An ML tree was then produced using the best fit model for the protein sequence data (JTT), bootstrap analyses with 1000 replicates and condensation of branches with less than 50% support. The partial MCP predicted protein from an ATV isolate from Utah (Accession Number AY548312.1) was used as the out group. The ORF57r protein data was analyzed and it was determined that the bestfitting amino acid substitution model was the Le Gascuel 2008 model [26]. An ML tree was built using the best-fit model for the protein sequence data and Bootstrap analyses with 1000 replicates were performed, but branch condensation was unnecessary. The partial ORF 57r predicted protein from an ATV isolate from Utah (Accession Number ACB11425) was selected as the outgroup.
Results
BLAST searches revealed high homology at both loci amongst the majority of the UK isolates and an isolate of FV3 (Accession Number AY58484.1; Tables 1 and 2). Four isolates exhibited high homology with a Ranavirus designated as Chinese Giant Salamander Iridovirus (CGSIV; 27). Homologies with CGSIV occurred at one (BUK 2, BUK 3, RUK 13) or at both loci (RT 119) (Table 1).
| Isolate |
Species |
Location |
Gene |
% Homology |
Isolate |
Accession Number |
|
BUK 2 |
Bufo bufo |
Unknown |
MCP |
100% |
FV3 |
KJ175144.1 |
| ORF57r |
100% |
CGSI |
KF512820.1 |
|
BUK 3 |
Bufo bufo |
Unknown |
MCP |
100% |
FV3 |
KJ175144.1 |
| ORF57r |
100% |
CGSI |
KF512820.1 |
|
BUK 4 |
Bufo bufo |
Unknown |
MCP |
100% |
FV3 |
KJ175144.1 |
| ORF57r |
100% |
FV3 |
KJ175144.1 |
| OS 14 |
Alytes obstetricans |
Brighton, East Sussex |
MCP |
100% |
FV3 |
KJ175144.1 |
| ORF57r |
98% |
FV3 |
KJ175144.1 |
|
RT 5 |
Rana temporaria |
Herne Bay, Kent |
MCP |
100% |
FV3 |
KJ175144.1 |
| ORF57r |
99% |
FV3 |
KJ175144.1 |
|
RT 8 |
Rana temporaria |
Unknown |
MCP |
100% |
FV3 |
KJ175144.1 |
| ORF57r |
98% |
FV3 |
KJ175144.1 |
|
RT 80 |
Rana temporaria |
Brighton, East Sussex |
MCP |
100% |
FV3 |
KJ175144.1 |
| ORF57r |
98% |
FV3 |
KJ175144.1 |
|
RT 112 |
Ranatemporaria |
Unknown |
MCP |
100% |
FV3 |
KJ175144.1 |
| ORF57r |
99% |
FV3 |
KJ175144.1 |
|
RT 115 |
Rana temporaria |
Unknown |
MCP |
100% |
FV3 |
KJ175144.1 |
| ORF57r |
99% |
FV3 |
KJ175144.1 |
|
RT 119 |
Rana temporaria |
Plymouth, Devon |
MCP |
99% |
CGSI |
KF512820.1 |
| ORF57r |
99% |
CGSI |
KF512820.1 |
|
RT 122 |
Rana temporaria |
Wokingham, Berkshire |
MCP |
100% |
FV3 |
KJ175144.1 |
| ORF57r |
99% |
FV3 |
KJ175144.1 |
|
RT 123 |
Rana temporaria |
Wokingham, Berkshire |
MCP |
100% |
FV3 |
KJ175144.1 |
| ORF57r |
99% |
FV3 |
KJ175144.1 |
|
RT 126 |
Rana temporaria |
Southampton, Hampshire |
MCP |
100% |
FV3 |
KJ175144.1 |
| ORF57r |
99% |
FV3 |
KJ175144.1 |
|
RT 127 |
Ranatemporaria |
Wallington, Surrey |
MCP |
100% |
FV3 |
KJ175144.1 |
| ORF57r |
99% |
FV3 |
KJ175144.1 |
|
RT 128 |
Rana temporaria |
Wallington, Surrey |
MCP |
100% |
FV3 |
KJ175144.1 |
| ORF57r |
99% |
FV3 |
KJ175144.1 |
|
RT 130 |
Rana temporaria |
Preston, Lancashire |
MCP |
100% |
FV3 |
KJ175144.1 |
| ORF57r |
99% |
FV3 |
KJ175144.1 |
|
RT 131 |
Rana temporaria |
Preston, Lancashire |
MCP |
100% |
FV3 |
KJ175144.1 |
| ORF57r |
99% |
FV3 |
KJ175144.1 |
|
RT 132 |
Rana temporaria |
Preston, Lancashire |
MCP |
100% |
FV3 |
KJ175144.1 |
| ORF57r |
99% |
FV3 |
KJ175144.1 |
|
RT 133 |
Rana temporaria |
Preston, Lancashire |
MCP |
100% |
FV3 |
KJ175144.1 |
| ORF57r |
99% |
FV3 |
KJ175144.1 |
|
RT 134 |
Rana temporaria |
Preston, Lancashire |
MCP |
100% |
FV3 |
KJ175144.1 |
| ORF57r |
99% |
FV3 |
KJ175144.1 |
|
RT 137 |
Rana temporaria |
Preston, Lancashire |
MCP |
99% |
FV3 |
KJ175144.1 |
| ORF57r |
99% |
FV3 |
KJ175144.1 |
|
RUK 11 |
Rana temporaria |
Unknown |
MCP |
100% |
FV3 |
KJ175144.1 |
| ORF57r |
99% |
FV3 |
KJ175144.1 |
| RUK 13 |
Rana temporaria |
Unknown |
MCP |
100% |
FV3 |
KJ175144.1 |
| ORF57r |
100% |
CGSI |
KF512820.1 |
| TT 216* |
Rana temporaria |
Deal, Kent |
MCP |
100% |
FV3 |
KJ175144.1 |
| ORF57r |
98% |
FV3 |
KJ175144.1 |
Table 1: Isolate abbreviations, locations of origin, and BLAST analysis results from UK Ranavirus isolates from three different amphibian species.
* Isolated from a tadpole
FV3–Frog virus 3
CGST–Chinese Giant Salamander Iridovirus
The best ML tree based on concatenated DNA sequence data contained several multifurcating branches that were roughly associated with the geographic origins of the isolates that make up the branches (Figures 1 and 2). Branches did not appear to be affected by the species of origin of the isolates, but instead the mortality event and/or geographic location of origin. Historical isolates that were obtained in the 1990s formed one clade, intermediate between isolates consistently exhibiting FV3-like DNA sequences, and the isolate exhibiting strong homology with CGSIV.
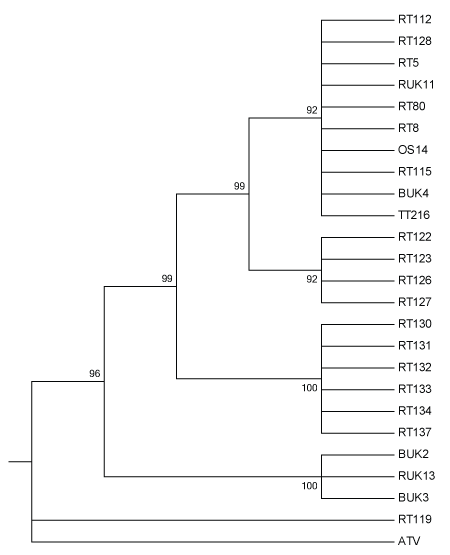
Figure 1: Maximum likelihood tree of 24 UK Ranavirus isolates using concatenated DNA sequences partial MCP and ORF57r genes using Kimura’s 2-Parameter model of nucleotide substitution and 1000 Bootstrap replicates. BUK=Isolate from B. bufo; RT and RUK=Isolate from R. temporaria; OS=Isolate from Alytes obstetricans; TT=Isolate from an R. temporaria tadpole.
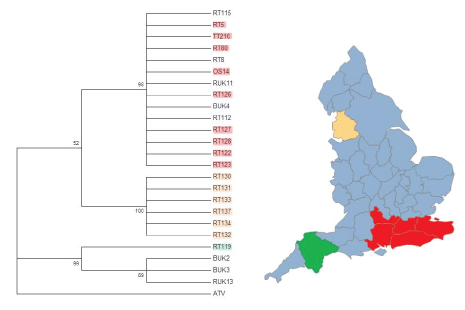
Figure 2: Phylogenetic analysis of partial ORF 57r DNA sequences from different locations around the UK genes using the Jukes-Cantor model of nucleotide substitution and 1000 Bootstrap replicates. BUK=Isolate from B. bufo; RT and RUK=Isolate from R. temporaria; OS=Isolate from Alytes obstetricans; TT=Isolate from an R. temporaria tadpole. Red highlighted isolates are from the southeast, orange/ peach highlighted isolates are from Lancashire, and green highlighted isolates are from Devon, and match the highlighted areas on the map of England. Isolates that are not highlighted do not have a known geographical origin. (Map created using SmartDraw 2017).
The tree based on the predicted protein sequences for the partial MCP loci included three multi-isolate clades (Figure 3), one composed of isolates from a single mortality event in Lancashire. Isolates containing CGSIV-like sequences (RT 119, BUK 2, BUK 3, RUK 13) formed another clade, despite being isolated from two different species, while the third clade contained a mix of modern and historical samples isolated from all three host species. Interestingly, RT 122 and RT 123 (both isolates from Berkshire) form a multifurcating branch. Additionally, RT 127, an isolate from Surrey, does not group with RT 128, another isolate from the same location, but forms a multifurcating branch with the outgroup, ATV.
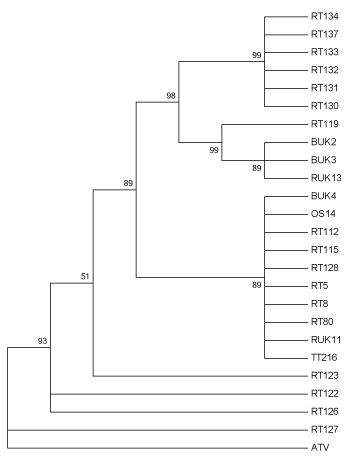
Figure 3: Maximum likelihood method tree of 24 UK Ranavirus isolates using the partial MCP sequence’s predicted protein sequence based on the JTT matrix-based protein substitution model and 1000 Bootstrap replicates. BUK=Isolate from B. bufo; RT and RUK=Isolate from R. temporaria; OS=Isolate from Alytes obstetricans; TT=Isolate from an R. temporaria tadpole.
Predicted protein produce of the partial ORF57r gene also returned three distinct clades (Figure 4). One clade was a multifurcating branch composed of 14 isolates from all three host species and including both modern and historical isolates. Again, isolates from a single mortality event in Lancashire formed a distinct clade, while the third clade included all isolates containing CGSIV-like DNA sequences.

Figure 4: Maximum likelihood method tree of 24 UK Ranavirus isolates using the partial ORF 57r sequence’s predicted protein sequence based on the Le Gascuel 2008 protein substitution model and 1000 Bootstrap replicates. BUK=Isolate from B. bufo; RT and RUK=Isolate from R. temporaria; OS=Isolate from Alytes obstetricans; TT=Isolate from an R. temporaria tadpole.
Discussion
BLAST searches revealed that the partial sequences of most isolates were highly homologous to an isolate of Frog virus 3 (FV3) that was obtained from a Northern leopard frog (Lithobates/Rana pipiens) that was captured in Northern Ontario, Canada by Morrison et al. ([27]; Table 1). This strain of FV3 is referred to by Morrison et al. [27] as SSME, and was found to be divergent from the previously published full genome sequences of FV3. These differences occurred at several loci, including some thought to be involved in virulence [27]. However, since we do not know the whole genome sequence we restrict our discussion to the interpretation of sequence similarity.
A smaller group of the English isolates partial sequences was most similar to an isolate, designated as Chinese Giant Salamander Iridovirus ([28]; Table 1). Phylogenetic analysis of other Ranaviruses derived from Chinese Giant Salamanders (Andrias davidianus) have shown that they are very closely aligned with the common midwife toad virus (CMTV) [15,29]. CMTV has been the causative agent of massive community level disease and declines in the Spanish Pyrenees [6].
Similarity to FV3 was not unexpected as FV3 is the type virus for the genus Ranavirus ([30]; Table 2). However, the high similarity of some isolates to an isolate of the CGSIV was unexpected as there have been no previous reports of similar isolates outside of China (Table 1). This, in combination with the fact that Ranavirus isolates from Chinese Giant Salamanders tend to be similar to CMTV, should be a cause for conservation concern for all English amphibian species. CGSIV appears to be highly virulent, having caused high mortality in both wild and captive Chinese giant salamanders (Andrias davidianus), which is a highly endangered species in China [2,31].
| Isolate |
Species |
Location |
Gene |
%
Homology |
|
BUK 2 |
Bufo bufo |
Unknown |
MCP |
100 |
| ORF57r |
94 |
|
BUK 3 |
Bufo bufo |
Unknown |
MCP |
100 |
| ORF57r |
94 |
|
BUK 4 |
Bufo bufo |
Unknown |
MCP |
100 |
| ORF57r |
100 |
|
OS 14 |
Alytes obstetricans |
Brighton, East Sussex |
MCP |
100 |
| ORF57r |
98 |
|
RT 5 |
Rana temporaria |
Herne Bay, Kent |
MCP |
100 |
| ORF57r |
99 |
|
RT 8 |
Ran atemporaria |
Unknown |
MCP |
100 |
| ORF57r |
98 |
|
RT 80 |
Rana temporaria |
Brighton, East Sussex |
MCP |
100 |
| ORF57r |
98 |
|
RT 112 |
Rana temporaria |
Unknown |
MCP |
100 |
| ORF57r |
98 |
|
RT 115 |
Rana temporaria |
Unknown |
MCP |
100 |
| ORF57r |
99 |
|
RT 119 |
Rana temporaria |
Plymouth, Devon |
MCP |
NR |
| ORF57r |
NR |
|
RT 122 |
Rana temporaria |
Wokingham, Berkshire |
MCP |
100 |
| ORF57r |
99 |
|
RT 123 |
Rana temporaria |
Wokingham, Berkshire |
MCP |
100 |
| ORF57r |
99 |
|
RT 126 |
Rana temporaria |
Southampton, Hampshire |
MCP |
100 |
| ORF57r |
99 |
|
RT 127 |
Rana temporaria |
Wallington, Surrey |
MCP |
99 |
| ORF57r |
99 |
|
RT 128 |
Rana temporaria |
Wallington, Surrey |
MCP |
100 |
| ORF57r |
99 |
|
RT 130 |
Rana temporaria |
Preston, Lancashire |
MCP |
100 |
| ORF57r |
99 |
|
RT 131 |
Rana temporaria |
Preston, Lancashire |
MCP |
100 |
| ORF57r |
99 |
|
RT 132 |
Rana temporaria |
Preston, Lancashire |
MCP |
100 |
| ORF57r |
99 |
|
RT 133 |
Rana temporaria |
Preston, Lancashire |
MCP |
100 |
| ORF57r |
99 |
|
RT 134 |
Rana temporaria |
Preston, Lancashire |
MCP |
100 |
| ORF57r |
99 |
|
RT 137 |
Rana temporaria |
Preston, Lancashire |
MCP |
99 |
| ORF57r |
99 |
|
RUK 11 |
Rana temporaria |
Unknown |
MCP |
100 |
| ORF57r |
99 |
|
RUK 13 |
Rana temporaria |
Unknown |
MCP |
100 |
| ORF57r |
94 |
|
TT 216* |
Rana temporaria |
Deal, Kent |
MCP |
100 |
| ORF57r |
98 |
|
|
|
|
|
Table 2: Comparison of isolates to Frog Virus 3 (Accession Number AY548484.1) which is the type virus for the genus Ranavirus. (NR=No Report, which means that no similarity/identify score was returned using BLAST comparing it to the type virus).
The phylogenetic tree found in Figure 1 has several multifurcating branches that are roughly associated with the geographic origin of the isolates (Figure 2). These branches also have high bootstrap support. The first clade at the top of the tree is made up of a branch with 10 isolates from three different host species. The isolates, RT 80 and OS 14, from R.temporaria and A. obstetricans, respectively, were obtained from the different species sampled during the same mortality event affecting a pond in Sussex in the southeast of England. This infection of an A. obstetricians adult may be the result of pathogen spillover from the disease outbreak seen in R. temporaria [11], as there is some evidence that the Ranavirus(es) present in England have developed some host specificity [7]. The isolate, TT 216, is from an R. temporaria tadpole that originated from a highly managed amphibian community in Kent, in the southeast of England [11]. Unfortunately, RUK 11, RT 112, RT 115, and BUK 4 are from unknown geographic origins, therefore we can only make an assertion that isolates within this clade are generally from the southeast of England. This clade is also made up of both historical (RUK and BUK) and contemporary isolates (OS, RT, TT), which is very interesting as the other historical isolates form a distinct clade further down the tree.
This second clade is another multifurcating branch, this time made up wholly of isolates derived from R. temporaria. These isolates originate from three separate mortality events in south-central England (Table 1). The presence of a clade restricted to R. temporaria is not surprising, because of the sampling bias towards R. temporaria as the targeted host [11,19].
The third clade is another multifurcating branch made up isolates solely from R. temporaria. Isolates RT 130-137 are all from the same mass mortality event in Lancashire (Figure 2). Since these isolates all are from the same mass mortality event, it is not surprising that they group together. It is possible that this clade represents a separate Ranavirus introduction event. It is geographically isolated and phylogenetically distinct and no other scientifically confirmed Ranavirus mortality events have been documented in the region (Figure 2). Due to the large distance between the southeast of England and Lancashire in the northwest, it is unlikely that this Rananvirus isolate was transported by amphibians during natural migrations.
The fourth clade from the top of the tree is also a multifucation; however, this one is made up solely of historical isolates. In this case, the branch is made up of isolates from both R. temporaria (RUK) and B. bufo (BUK). Since these isolates are from early in the timeline of emergence of Ranaviruses in England, it is possible that sequence divergence has not yet become established at the two loci examined, although genetic divergence in these lineages divergence is suspected because of differences reported in the Ranavirus biology [7]. Alternatively it could simply be that these two loci are not involved in host specificity. This same branching pattern is seen in Price et al. [15] who used different regions of the genome and longer concatenated sequences.
The isolate RT 119, formed an unexpected multifurcating branch with the outgroup, ATV (Figure 1). RT 119 was also highly homologous with an isolate of CGSIV at both loci examined (Table 1). Since, RT 119 is from the west of England (Devon), it is possible that it originated from a separate introduction event, perhaps from Asia, as no other Ranavirus associated mortality events have been described in western England. Also, due to the large geographical distance between the southeast of England and the southwest, it is unlikely that the Ranavirus was transported in either direction through the natural movements of any affected species.
The trees that were made for the predicted protein sequences at both loci suggest that Ranaviruses have been introduced into the UK, specifically, England, at least three times. It is likely that BUK 2, BUK 3, and RUK 13 all are from the same introduction event as they show the same pattern of homology with both FV3 and CGSI in all four trees.
Price et al. [15] combined citizen science data with genetic information from seven UK isolates to suggest at least two introductions of Ranavirus into the United Kingdom. They used a comparatively small portion of the full Ranavirus genome (about 2267 bp or less than 2%). Our more extensive sampling extends Price et al. [15] conclusions, by suggesting that there have likely been at least three introductions of FV3-like Ranaviruses into England alone. This can be seen from the three major branching points in Figures 1-4. Our data also supports Price et al. [15] finding that humans have enhanced the spread the disease-causing agent across the UK, especially in southeast England. The geographic clustering of the isolates suggests that a combination of pond to pond animal movement and human movement of animals has contributed to the spread of the different strains of the infectious disease into adjoining regions [15]. Human enhanced spread has been noted in the emergence of other Ranaviruses (e.g. ATV) [32] and it is highly likely to be the origins of the Devon and Lancashire isolates (Figure 2).
Based on the different phylogenetic analyses of two partial genes, both with them concatenated, and using their predicted protein products, we show that Ranaviruses have been introduced into the UK, specifically, England, at least three times. High homology of isolates with Chinese Giant Salamander Iridovirus should be cause for great conservation concern as it is very similar to common midwife toad virus, which has decimated amphibian communities in the Spanish Pyrenees [6]. Further investigations that examine larger portions of the viral genome, or even full genomes, are needed to fully understand the evolutionary history of Ranaviruses in the UK, as well as the host-strain associations that are known to exist [7].
Acknowledgments
We would like to thank Andrew Cunningham for the provision of the BUK and RUK isolates and Thomas Waltzek for his valuable advice on phylogenetic reconstruction methods. We would also like to thank James Jancovich, John C. George, and several anonymous reviewers for helpful comments on earlier versions of this manuscript. This work was supported by a PhD studentship awarded to ALJD by Queen Mary University of London, an Overseas Research Studentship, as well as one provided by the National Science and Engineering Research Council of Canada. Additional support was provided by a Convocation Research Trust Award, Amphibian Conservation Research Trust Student Research Grant, and a British Wildlife Health Association Grant to ALJD and an RCUK Fellowship awarded to TWJG.