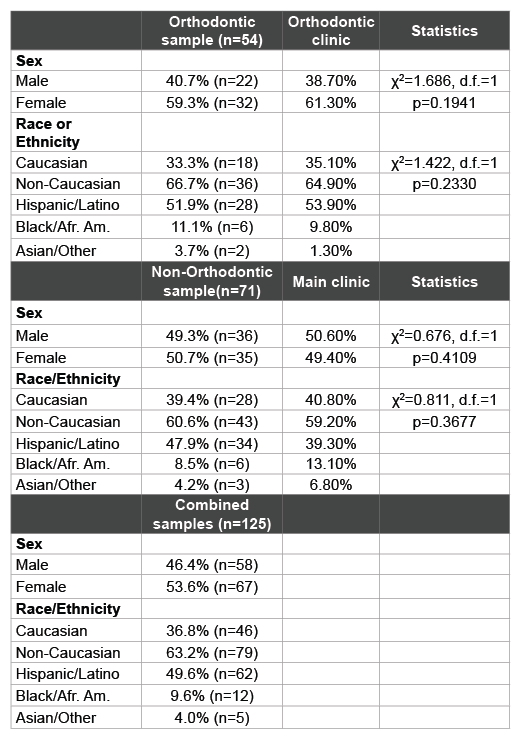
Table 1: Patient sample and clinic characteristics


David Jolley Kaylee Wonder Ecsile Chang Karl Kingsley*
University of Nevada, Las Vegas, School of Dental Medicine, Nevada, USA*Corresponding author: Karl Kingsley, University of Nevada, Las Vegas, School of Dental Medicine, Nevada, USA, Tel: +702-774-2625; E-mail: Karl.Kingsley@unlv.edu
Background: Changes in the oral microbial flora are commonplace during orthodontic therapy, although some evidence suggests these alterations may extend for some time after. Although many studies have screened for changes in cariogenic pathogen levels, more evidence is accumulating to demonstrate significant changes among periodontal pathogens within these patients. Although several studies at this predominantly low-income, dental school-based Orthodontic clinic have screened for cariogenic pathogens-none to date have provided multiorganism screening for periodontal pathogens.
Objective: This goal of this study was to complete a retrospective, cross-sectional study of saliva samples to screen for Fusobacterium nucleatum, Treponema denticola, and Porphyromonas gingivalis among orthodontic and non-orthodontic patients (n=125).
Methods: Using previously collected saliva samples, DNA was isolated and screened using PCR using primers specific for each pathogen of interest. Differences in prevalence between groups (Orthodontic, non-Orthodontic) were measured using Chi-square analysis.
Results: This analysis revealed the presence of these pathogens in nearly half of orthodontic patient samples and more than half of nonOrthodontic samples. These data also demonstrated females exhibited greater prevalence than males, while the overall prevalence among nonorthodontic samples was greater. This may be associated with higher average age, larger body mass index (BMI) and greater periodontal pocked depth (PPD) and decayed-missing-filled teeth (DMFT) scores.
Conclusion: These findings suggest the strong need to plan and implement a prospective study to determine the baseline prevalence of these pathogens among this patient population as they begin orthodontic therapy and how these levels change over time. This may provide more relevant clinical information for oral health scientists and local epidemiologists to determine the most vulnerable populations, as well as the best methods and timing for interventions to prevent poor oral health outcomes and long-term consequences associated with periodontal disease.
Periodontal pathogen; Orthodontics; Saliva screening
Although many studies of oral microbial changes during Orthodontic therapy have necessarily focused on cariogenic pathogens [1,2], fewer studies have closely examined the changes to other oral flora, including periodontal pathogens [3,4]. Studies have demonstrated that orthodontic treatment alters the oral microbiome and can both directly and indirectly alter the oral microbial composition, thereby dramatically increasing the potential for both cariogenic and periodontal disease [5-7]. Recent evidence has suggested that microbial alterations during orthodontic treatment may outlast the duration of therapy and influence long-term oral health outcomes [8-10].
Many studies have demonstrated normal, baseline ranges for levels of potential periodontal pathogens in the oral biofilm and subgingival crevices, which may trigger disease if homeostasis is disrupted [11,12]. These pathogens, include Fusobacterium nucleatum (FN), Treponema denticola (TD), and Porphyromonas gingivalis (PG)-the major etiologic agent implicated in chronic and persistent periodontitis [12,13]. Although modern materials and Orthodontic techniques have improved oral health outcomes in recent years, all current treatments are associated with increased levels of periodontal pathogen levels to some degree in many patients [14,15].
New diagnostic methods involving salivary biomarkers have improved the ability to monitor oral and periodontal diseases in recent years [16,17]. These advances facilitate studies investigating salivary screening for oral microbial changes during Orthodontic treatment [18,19]. In fact, studies from this school have utilized salivary biomarkers to screen for cariogenic pathogen changes among Orthodontic clinic patients-although no large-scale screening for periodontal pathogen levels has yet been attempted within this patient population [20-22].
Our studies have informed us that oral health status among orthodontic patients, particularly at this dental school-based clinic, may be of particular concern due to the large number of low-income and Minority patients who may face greater barriers and challenges to receive high quality healthcare [23,24]. The higher prevalence of these cariogenic pathogens, combined with increased barriers and lowered access to care, may explain some of these observations -although the full spectrum of changes within the oral microbial flora remains incomplete. These data serve as the basis for the current study objective, which is to screen orthodontic and non-orthodontic patients from this dental school patient clinic and determine the relative prevalence of specific periodontal pathogens, such as FN, TD and PG.
The protocol submission “Retrospective investigation of oral microbes from the UNLV-SDM patient population” (OPRS#762911-1) was approved by the UNLV Biomedical IRB on August 3, 2015. Saliva samples were originally collected and appropriately archived from a convenience sample of eligible patients. Exclusion criteria included patients that chose not to participate, patients aged seven or younger, and adult patients with oral cancer. The approval for the original study “The prevalence of oral microbes in saliva from the UNLV School of Dental medicine pediatric and adult clinical population” was granted in May 2013 by the Office of Research Integrity and Protection of Research (Human) Subjects (OPRS#1305-4466M). This project will retrospectively examine a number of these samples (n=125).
Although this is a retrospective study, the original protocol involved in-clinic saliva collection. As samples were collected, each was assigned a unique, non-duplicated number generated at random to preserve patient confidentiality and prevent research bias.
In addition to the saliva collection, some demographic data was also obtained from each patient. This included the sex, age and self-reported race or ethnicity, as well as some biometric data, including body mass index (BMI) parameters such as height and weight, as well as some clinic observations for decayed, missing, or filled teeth (DMFT), and depth of periodontal pockets (PPD).
Following the saliva collection, each sample was kept cool (using ice) until processed. All samples were processed using a standard aliquot (500 µL) and the Genomic Prep DNA isolation kit from Amersham Biosciences (Buckinghamshire, UK) as previously described [20-22]. The quality and quantity of DNA was determined using absorbance readings of 260/280 nm.
To screen for the pathogen of interest (FN, TD or PG), a standard amount of isolated DNA was utilized using the exACTGene complete PCR kit from Fisher Scientific (Fair Lawn, NJ, USA) and primers for TD, FN, PG and the human enzyme (control) glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which were made by SeqWright (Houston, Texas, USA)
TD primer (forward); 5’-TAATACCGAATGTGCTCATTTACAT-3’
TD primer (reverse); 5’-CTGCCATATCTCTTGTCATTGCTCTT-3’
FN primer (forward); 5’-CGCAGAAGGTGAAAGTCCTGTAT-3’
FN primer (reverse); 5’-TGGTCCTCACTGATTCACACAGA-3’
PG primer (forward); 5’-TACCCATCGTCGCCTTGGT-3’
PG primer (reverse); 5’-CGGACTAAAACCGCATACACTTG-3’
GAPDH primer (forward); 5’-ATCTTCCAGGAGCGAGATCC -3’
GAPDH primer (reverse); 5’-ACCACTGACACGTTGGCAGT-3’
Each PCR reaction had an identical setup, using a standardized amount of DNA (1 µg). The basic parameters were denaturation at 94°C for three minutes, then 30 amplification cycles that consisted of denaturation at 94°C for 20 seconds , annealing at varying temperatures (based upon the primer sequence) for 60 seconds, extension at 72°C for 30 seconds with a final extension at 72°C for five minutes. Results were visualized using a Kodak Gel Logic 100 Imaging System and 1D Image Analysis Software (Eastman Kodak: Rochester, New York, USA) following gel electrophoresis using Reliant agarose gels (Lonza: Rockland, Maine, USA) and UV illumination using ethidium-bromide.
A DNA standard was creating using an existing human cell line, HGF-1 to determine the minimum cell number needed for relative endpoint or RE-PCR comparison. This DNA allowed for the determination of the PCR conditions, also known as the minimum cycle threshold or CT that is the minimum number of PCR cycles needed to visualize a known quantity of DNA amplified by PCR and the maximum cycle saturation point or CS, as described in previous work [20-22]. Using this standard and method, CT was determined to be twenty cycles (C20) with saturation observed at thirty five cycles (C35).
Porphyromonas gingivalis or PG was purchased from ATCC (FDC-381; Manassas, VA), as previous described [20,21]. Using over night growth suspensions, absorbance readings at 650 nm with an optical density (OD) reading of 0. 8 were found to approximate 107 CFU/mL. Dilutions of this suspension were made to yield cell number of 5.0 × 106 , 105 , 104 and 103 CFU/mL, which represent salivary microbial concentrations that correspond to disease risk ranging from 106 CFU/mL representing very high riskto 103 CFU/mL which represents normal or average risk. Threshold or CT for PG was found to require twenty five cycles (C25) and saturation was found to be forty five cycles (C45). Combining the data from the GAPD and PG experiments, CT was C20 and C25, respectively, while CS was C35 and C45, respectively [20,25,26]. Based upon this information, RE-PCR was performing using an intermediate cycle within those ranges at C30, which was in between the detection (CT) and saturation (CS) limits for both organisms.
The sample size was initially determined using the lower estimated DNA recovery rate from the DNA extraction kit (90%) to provide a minimum expected difference of 0. 10. To obtain statistical power of p=0. 80 and significance level, α=0. 05-a minimum sample size (n=50) was necessary [27]. Chi square analysis was used to determine any differences in categorical data regarding patient demographics (Sex, Race/Ethnicity), as well as any differences in TD, PG or FN between groups (based on Sex, Race/Ethnicity).
Saliva samples were grouped based upon the clinic from which the patients were originally recruited, which included the orthodontic clinic and (non-orthodontic) Main Patient clinics (Table 1). The orthodontic sample reflected an overall distribution similar to the overall distribution within this clinic population. For example, the samples derived from patients in the Orthodontic clinic (n=54) contained more females (59.3%) than males (40.7%), which was roughly similar to their overall distribution within the overall Orthodontic clinic (p=0.1941). Moreover, the percentage of samples from minority patients (66.7%) reflected approximately the same percentages within the orthodontic clinic overall (64.9%) and was not statistically significant (p=0.2330). In addition, the vast majority of these minority patients self-identified as Hispanic (n=28/36=77.8%).

Table 1: Patient sample and clinic characteristics
The samples collected from the non-orthodontic or Main patient clinic were nearly equally distributed among females (50.7%) and males (49.3%), which was similar to their percentages within the overall main clinic population (49.4%, 50.6%, p=0.4109). The majority of patients identified themselves as racial or ethnic minorities (60.6%), which was also similar to the overall clinic patient composition (59.2%, p=0.3677). As with the Orthodontic clinic samples, the overwhelming majority of these minority patients were Hispanic (n=34/43 or 79.1%).
These corresponding patient samples were then subjected to the DNA isolation procedure prior to screening and analysis (Table 2). These data revealed a recovery rate of 98.4% (n=123/125), comparable to previous studies [20,21,28,29]. DNA concentrations averaged 474.5 ng/uL, which ranged from 578.5 ng/uL in the orthodontic samples to 393.2 ng/uL in non-orthodontic patient samples. Purity of DNA ranged between 1.61 and 2.0, allowing for the screening by PCR which demonstrated the presence of both human (GAPDH) and bacterial (16S rRNA) DNA.
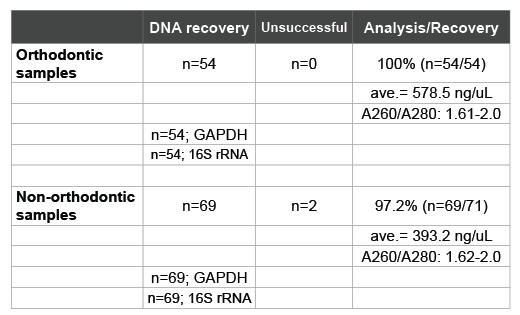
Table 2: Recovery and isolation of DNA
As described in the Materials and Methods section, DNA standards were generated to find the threshold and saturation PCR cycles (CT, CS) generally used to compare relative starting DNA concentrations in relative endpoint PCR (Figure 1A). Using these standards and methods, CT for GAPDH was observed at C20 and for PG at C25, with the corresponding CS at C35 and C45, respectively, RE-PCR was subsequently completed at C30, which was higher than the lower detection limit (CT), but still below the limits of saturation (C35-C45) for both.
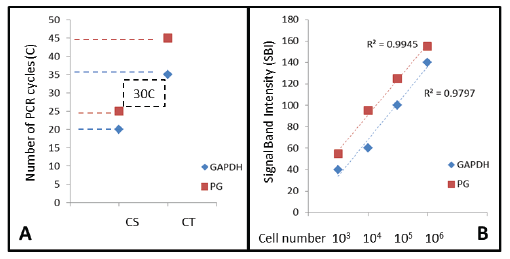
Figure 1: DNA standards and quantitative analysis PCR Cycle Threshold (CT) or detection limit and Cycle Saturation (CS) were determined for human (GAPDH) and bacterial (PG) cells, revealing the optimal screening cycle between C25 and C35. PCR signal band intensity (SBI) was strongly correlated with starting cell number (R2 >0.97) at C30, which will allow for an approximation of starting cell number from the saliva samples screened.PG (P. gingivalis); GAPDH (glyceraldehyde-3-phosphate dehydrogenase)
Dilutions of standardized cell numbers 106 , 105 , 104 and 103 cells/ mL (human) or CFU/mL (bacteria) were processed accordingly. These numbers approximate research demonstrating salivary microbial concentrations and disease risk associations [20,25,26]:
106 CFU/mL indicates very high risk;
105 CFU/mL indicates high risk;
104 CFU/mL indicates moderate risk;
<103 CFU/mL indicates normal or average risk
These serial dilutions were prepared to establish PCR standard curves for both GAPDH and PG (Figure 1B). These data indicate that signal band intensity (SBI) at cycle 30 (C30) is nearly perfectly correlated with the starting cell number for both PG (R2 =0.9945) and GAPDH (R2 =0.9797).
Following DNA isolation, all samples were screened for the presence of F. nucleatum (FN), T. denticola (TD) and P. gingivalis (PG) at levels at or above pre-determined disease-risk levels (>103 CFU/mL) as described by previous saliva-based PCR screening studies (Figure 2) [20-22]. These data revealed that FN, TD and PG were present at or above these predetermined levels in 52%, 41.6% and 48% of all samples, respectively. More specifically the prevalence of FN, TD and PG within the Orthodontic samples (46.3%, 38.9%, 44.4%) was significantly lower than the control, non-Orthodontic samples (56.3%, 43.7%, 50.7%)

Figure 2: PCR screening of DNA isolated from saliva Using previously established DNA standards to determine the PCR cycle threshold detection standards for >104 CFU/mL, nearly half of all samples were found to harbor FN, PG and TD. More detailed analysis revealed the Orthodontic samples had significantly lower prevalence of FN (p<0.01) and PG (p<0.05), as well as lower prevalence of TD (p=0.07) than non-Orthodontic samples. FN (F. nucleatum), TD (T. denticola); PG (P.gingivalis); χ2 (Chi squared), d.f. (degrees of freedom)
To determine if the differences in prevalence of FN, TD and PG between the Orthodontic and non-Orthodontic clinic samples were due to other factors, more detailed analyses were performed to evaluate any possible influence by Sex/Gender (Figure 3). Although a general pattern of significantly lower periodontal pathogen prevalence was found among all the Orthodontic samples, only FN prevalence among Male Orthodontic patients, specifically, was significantly higher than expected (p<0.01). Moreover, although a higher prevalence of periodontal pathogens was observed in the non-orthodontic (control) samples-a gender / sex specific pattern was also evident with females exhibiting significantly higher levels of all periodontal pathogens than males, but proportionally much higher levels of FN and PG (p<0.01). However, no significant differences were observed between Racial or Ethnic categories.
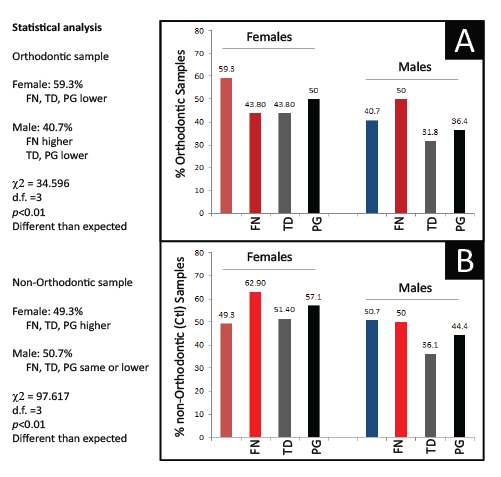
Figure 3: Analysis of PCR screening by sex Sorting of orthodontic samples into Females and Males revealed an overall pattern of lower pathogen prevalence except among a significantly higher proportion of Male orthodontic patients (p<0.01). The analysis of non-orthodontic (control) samples also revealed a sex-specific pattern with significantly higher proportions of Females exhibiting pathogen prevalence than Males (p<0.01).FN (F. nucleatum), TD (T. denticola); PG (P.gingivalis); χ2 (Chi squared), d.f. (degrees of freedom)
The additional demographic and health data from each patient sample was also analyzed and reviewed (Table 3). This information included patient age, body mass index or BMI, periodontal pocket depth (PPD) and decayed, missing, and filled teeth (DMFT) score, which were grouped by clinic (Orthodontic or Main clinic) and then sorted by gender and ethnicity. This analysis revealed that the average age of patients from the orthodontic sample (24.4 years) was significantly lower than those from the Non-orthodontic sample (28.3 years). Although no striking differences were found among the ages of males and females or minorities and non-minorities from the Orthodontic sample, there were much larger differences from the non-Orthodontic sample. In addition, average BMI was also significantly higher within the non-orthodontic sample (29.3) than the Orthodontic sample (25.7) with only minor differences observed between genders or by race and ethnicity.
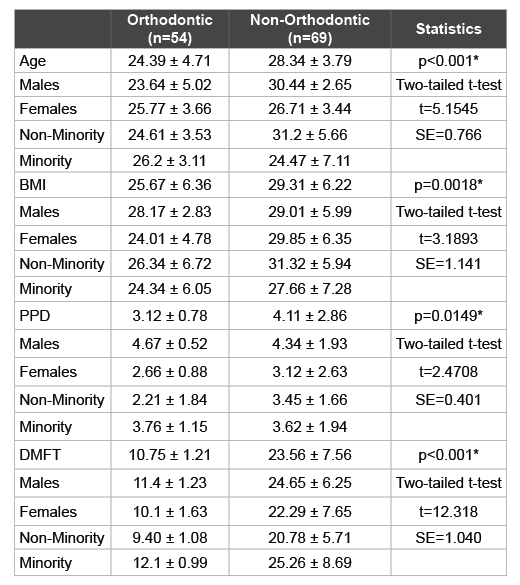
Table 3: Analysis of study sample demographic and health parameters
*denotes statistical significance (p<0.05); BMI (Body Mass Index); PPD
(Periodontal Pocked Depth); DMFT (Decayed-Missing-Filled Teeth); t
(critical value of t-statistic for two-tailed test); SE (Standard Error)
Interestingly, PPD was much greater within the non-Orthodontic samples (4.11) compared with the Orthodontic samples (3.12), which varied widely. More specifically, males within the Orthodontic sample had much greater PPD (4.67) than females (2.66) while Minorities exhibited greater PPD (3.67) than Whites (2.21). These differences were not observed within the non-Orthodontic sample. As expected, DMFT score varied significantly with lower scores among the Orthodontic sample (10.75) compared with the non-Orthodontic samples (23.56) and with higher DMFT scores among Minorities from either clinic.
Finally, an analysis was performed to compare the PCR screening results of this study with the health parameters concurrently collected (Table 4). As most of the health parameters were significantly different between the orthodontic and non-orthodontic samples, this analysis may reveal relationships between these additional variables and the results of the PCR screening. For example, although there were significant differences between the orthodontic and non-orthodontic samples with respect to FN and PG prevalence, these differences did not appear to be significantly related to the overall age or the average BMI from these two groups. However, there were some significant associations between the clinical oral health parameters measured and periodontal pathogen detection, such as the association between PPD and TD prevalence, as well as the overall DMFT averages and FN, TD and PG prevalence.
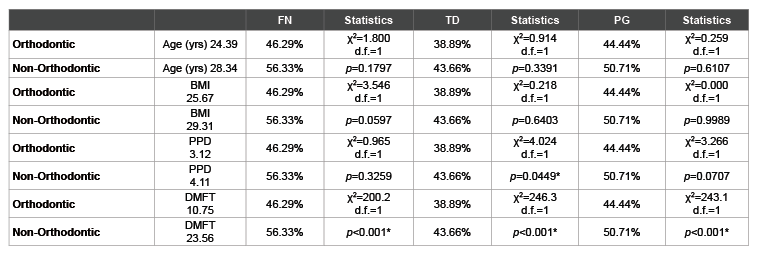
Table 4: Analysis of health parameters and screening results
*denotes statistical significance (p<0.05); FN (F. nucleatum), TD (T. denticola); PG (P.gingivalis); yrs (Years); BMI (Body Mass Index); PPD (Periodontal
Pocked Depth); DMFT (Decayed-Missing-Filled Teeth); χ2
(Chi squared), d.f. (degrees of freedom)
The objective of this study was to determine the oral microbial burden of specific periodontal pathogens among orthodontic patients for comparison with non-orthodontic controls. As recent evidence has suggested, many studies of changes to the oral microbial flora among Orthodontic patients have focused largely on cariogenic pathogens, while fewer studies have examined the potential changes associated with specific periodontal pathogens, such as T. denticola, F. nucleatum and P. gingivalis -particularly among adult patients. Although this study utilized previously collected saliva samples and not the more specific, and clinically relevant gingival crevicular fluid (GCF), these methods are consistent with previous retrospective studies from this patient population [20-22] and other recently published works that utilize existing saliva repositories [7,16,18]. The outcomes of this study clearly demonstrated observable differences found between samples from Orthodontic and non-Orthodontic patients.
Unlike previous studies of this orthodontic patient clinic, which demonstrated much higher prevalence of oral cariogenic pathogens [20,22], the results of this study found significantly lower levels of periodontal pathogens within this patient sample compared with the main patient clinic. One potential explanation for these observations could be the disproportionately high percentage of very low income, first-time dental visits among the main clinic population, which may be considerably different from Orthodontic patients that have been through several screening, hygiene, and follow-up appointments [20,22-24]. In addition, the current study sample size (n=125) is larger than any of the previous studies evaluated (ranging from n=52 to n=75) which may also have influenced these findings. Interestingly, the most recent study from this school found mostly cariogenic and one periodontal pathogen (PG) in nearly half of the Orthodontic samples, which roughly compares with the results of the current study [22]. The non-orthodontic samples from that previously study, however demonstrated only about 25% harbored PG at or above disease risk levels, which is far lower than the findings of this current study-suggesting that more research will be needed to further elucidate the disparate nature of these results.
This study has several limitations that must also be considered when evaluating the results and conclusions. The retrospective study design may have significantly affected the results through selection bias of the recruitment team or other confounding factors, such as patient participation or self-selection bias [20-22]. In addition, collection of these samples at only one patient visit and time point suggests that no temporal conclusions can be made regarding the observations in periodontal pathogen prevalence from this type of cross sectional study. Because only limited information regarding each patient sample was available, no attempt was made to standardize the amount of time a patient was in treatment within the Orthodontic treatment, which may further limit the overall conclusions that can be drawn from these results.
Because of the significant differences observed between these groups, future studies of this population should be prospective in nature to gather clinical samples from each individual patient before treatment has begun and corresponding samples taken at various time points following Orthodontic bracket placement. This may help to disentangle some of these possible confounding variables [30]. This type of prospective, multitime point analysis would also allow for the incorporation of orthodontic treatment duration as another potential variable of interest for analysis [31]. Additional data of interest, including assessment of clinical hygiene and surveys of oral hygiene and dietary patterns could also be concurrently collected to more accurately determine the nature of differences within and between these groups over time [3,32]. Finally, a prospective study would allow clinical samples from saliva to be compared with samples derived from the gingival crevicular fluid (GCF), as well as samples taken directly from various sites from the tooth surface or sites proximal to the Orthodontic brackets, which would allow for more robust analysis of these observed phenomenon [33,34].
Despite these limitations, these findings are among the first to describe in detail the prevalence of periodontal pathogens among this patient population and the potential association with various health and demographic factors. These findings suggest the strong need to plan and implement a prospective study to determine the baseline prevalence of PG, FN and TD among these patients as they begin Orthodontic therapy and how these levels may change over time. This may provide more relevant clinical information for oral health scientists and local epidemiologists to determine the most vulnerable populations, as well as the best methods and timing for interventions to prevent poor oral health outcomes and long-term consequences associated with acute periodontal disease.
Download Provisional PDF Here
Article Type: Research Article
Citation: Jolley D, Wonder K, Chang E, Kingsley K (2016) Oral Microbial Prevalence of Periodontal Pathogens among Orthodontic Patients. Int J Dent Oral Health 2(6): doi http://dx.doi.org/10.16966/2378-7090.210
Copyright: © 2016 Jolley D, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Publication history:
All Sci Forschen Journals are Open Access