Full Text
Stanislav I Svetlov 1,2,3* Victor Prima 3 Zhiqun Zhang 2 Kenneth C Curley 4 Firas Kobeissy 2 Ahmed Moghieb 2 Kevin KW Wang 111Banyan Laboratories, Banyan Biomarkers, Inc, Alachua, FL USA
2Departments of Psychiatry University of Florida, Gainesville, FL, USA
3Department of Medicine, University of Florida, Gainesville, FL, USA
4U.S. Army Medical Research and Materiel Command (USA MRMC), Fort Detrick, MD, USA
*Corresponding author: Stanislav I Svetlov, MD, PhD, Research Associate Professor, 100
Newell Drive Gainesville, FL 32611, USA, Tel: 352-359-1612; E-mail: ssvetlov@ufl.edu
Article Information
Article Type: Research Article
Citation: Svetlov SI, Prima V, Zhang Z , Curley KC, Kobeissy F, et al. (2015) Acute and Subacute Differential Gene Expression in Rat Midbrain Following Blast Exposure Compared to Mechanical Brain Trauma. J Neurol Neurobiol, Volume1.1: http://dx.doi.org/10.16966/2379-7150.103
Copyright: © 2015 Svetlov SI, et al. This is an openaccess article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Publication history:
Abstract
In this study, we employed two models of traumatic brain injury (TBI) in rats: head-directed blast overpressure exposure (OBI) and mechanical controlled cortical impact (CCI). Two groups of animals served as controls: naïve rats and rats exposed to blast noise. A genomics neurosystems biology approach was used to analyze differential gene expression between experimental groups. Midbrain RNA was isolated 24 hours (acute) and 7 days (subacute) after traumatic insult, probed with Affymetrix array containing 31,100 gene sequences, and quantified by Agilent Technologies. Differentially expressed genes were grouped into functional clusters and the significance of gene expression changes at 24 h and 7 days was further assessed using parametric and nonparametric t- test with Welch correction. Then, gene interactions in pathological pathways following blast exposures vs. mechanical impact were created and analysed using neurosystems biology tools.
According to ANOVA (p<0.05), there was a significant difference in expression of 994 genes within blast exposure groups (vs. control) and in 1532 genes within CCI group (vs. control) with an overall overlap of 579 genes. Parametric and nonparametric t-tests revealed significant differences in temporal profile of changes in various genes at 24 hours vs. 7 days in blast exposed animals compared to CCI. Specifically, genes involved in neural development and repair, such as ROBO1 and NEDD4 were up-regulated in CCI, while they were shown to be down-regulated following blast exposure.
These results demonstrate several differences as well as overlaps in the expression of genes between the two brain insults. This will help to reveal specific molecular signatures of each brain insult and assist in developing diagnostics of chronic posttraumatic encephalopathy.
Keywords
Blast brain injury; Controlled cortical impact; Gene expression; Neurorepair; Genomics; Neurosystems biology
Introduction
Despite remarkable progress in understanding traumatic brain injury (TBI), particularly combat related TBI, the diverse pathological pathways and long-term outcome remain to be revealed and delineated [1]. Specifically, the question has to be addressed is to what degree the mechanisms of blast-induced TBI are similar to to mechanical impact type of brain injury, including concussion, and what may be the differences. It is especially important to elucidate the molecular dynamics of changes at different levels-of molecular regulation; this would include gene expression studies (mRNA) and its regulation (miRNA), proteomics and its associated area of metabolomics.
Several studies from our lab and others have been conducted to establish a comprehensive picture of pathology underlying traumatic brain injury, including genomic and proteomic signatures [2-4]. However, there are reports available that analyse side-by-side the genomic changes occurring after blast exposures compared to mechanical insult, such as controlled cortical impact (CCI) or closed head injuries. Also, it appears to be critical to employ neurosystems biology tools to reveal and visualize gene interactions, molecular functions and biological processes involved in such brain insults, as well as to decipher the overlaps or differences in pathways underlying non-penetrating blast-induced injury and mechanical CCI damage.
The main objective of this study was to apply an advanced genomics coupled with neurosystems biology tools to compare altered gene expression patterns of moderate blast exposure with brain injury produced by a mechanical Controlled Cortical Impact (CCI) at different times after challenge. Thus, for this purpose, gene ontology analysis including molecular function and biological process were employed for the differential gene identified; in addition, a a global approach of gene interaction was to see how the genes are related along with a targeted approach to identify downstream targets are being involved due to this interaction.. This allowed us to reveal time-dependent similarities and injury-specific differences between blast and CCI brain insults and reveal critical components of neuroinflammation, neuromediator and neurorepair components.
Materials and Methods
Animal exposure to a controlled blast wave: Modeling blast overpressure exposure was achieved by variable positioning of the target vs. blast generator described in detail previously [5]. The blast pressure data was acquired using PCB piezoelectric blast pressure transducers and LabView 8.2 software. Adult male rats (Sprague-Dawley, Harlan, Indianapolis, IN, USA) were anesthetized with 4% isoflurane in a carrier gas of 1:1 O2/N2O (4 min) [5]. After reaching a deep plane of anesthesia, they were placed into a holder exposing at the distance 5 cm below the exit nozzle of the shock tube. Rats were positioned directly on the shock tube. axis to expose them to the ‘composite’ blast including the compressed air jet with a peak overpressure of 52 psi, 10 msec duration. The control group of animals underwent anesthesia, placed in the cage aside shock tube and blast was produced thus exposing animals to the noise only.
Controlled Cortical Impact (CCI)
A controlled cortical impact (CCI) procedure was used to model mechanical TBI on rats essentially as previously described [6]. Briefly, rats were anesthetized with 4% isoflurane as above and maintained in 2.5% isoflurane. Core body temperature was monitored continuously and maintained at 37 ± 1°C. Animals were mounted in a stereotactic frame in a prone position and secured by ear and incisor bars. Following a midline cranial incision and reflection of the soft tissues, a unilateral (ipsilateral to site of impact) craniotomy was performed adjacent to the central suture, midway between bregma and lambda. The dura mater was kept intact over the cortex. Brain trauma was produced by impacting the right (ipsilateral) cortex with a 4 mm diameter impact or tip attached to electromagnetic driver at a velocity of 3.5 m/s with a 2.5 mm compression and 200 ms dwell time.
Appropriate pre- and post-injury management for both blast and CCI was preformed to minimize pain and discomfort and to insure compliance with guidelines set forth by the University of Florida, Institutional Animal Care and Use Committee and the National Institutes of Health guidelines detailed in the Guide for the Care and Use of Laboratory Animals. In addition, research was conducted in compliance with the Animal Welfare Act and other federal statutes and regulations relating to animals and experiments involving animals and adhered to principles stated in the “Guide for the Care and Use of Laboratory Animals, NRC Publication, 1996 edition.”
Brain tissue collection: At 24 hours or 7 days post-TBI, animals were euthanized; brain tissues were dissected with Brain Slicer device from the midbrain area (4-5 mm thick) according to the scheme (Figure 1). The central part of section (from about -5 mm to -9 mm posterior of Bregma) was cleared of surrounding cortex, pituitary and other tissues. It was cut in 2 halves and snap-frozen for proteomics (L), or preserved in RNALater (Ambion, Austin, TX) for RNA studies.
RNA isolation, quality characterization and validation were assured (Agilent Bioanalyzer, Santa Clara, CA) and genomic profiling was conducted according QC/QA guided procedures using Affimetrix GeneChip array at Assuragen, Inc. (Austin, TX). The raw fluorescent data were collected after hybridization and array scanning using Affymetrix GCOS v1.3. The data were further normalized and calculated using the Affymetrix MAS 5.0 algorithm. The summarized values were analyzed using one-way ANOVA and differences between paired naïve, noise control, and TBI groups were further determined by parametric and non-parametric t-tests with Welch correction (GraphPad, Inc.). Pathway maps were created using Systems Biology software PathwayStudio (Ariadne Genomics, Rockville, MD). Other Analysis Software: Partek Genomic Solutions 6.2 (Partek Inc., St. Charles, MO); Matlab 7 (Mathworks Inc., MA).
Neurosystems Biology Analysis
The genomics microarray differential expression of the CCI , blast injury cohorts vs. their respective controls were further analyzed using a bioinformatics systems biology approach to assess the altered pathway(s) relevant to differential specific brain injury altered genes and their its contribution to the development of either CCI and/or blast injury. PathwayStudio software (v 10; Ariadne Genomics, Rockville, MD, USA) was applied for the neurosystems biology analysis. This software helps to interpret biological meaning from differential gene expression, build and analyze pathways, and identify altered cellular processes and molecular functions involved.
Results and Discussion
According to ANOVA (p<0.05), there was a significant difference in expression of 994 genes within blast exposure groups (vs. control) and in 1532 genes within CCI group (vs. control) with overall overlap of 579 genes. Parametric and nonparametric t-tests revealed significant differences in temporal profile of changes in various genes at 24 hours vs. 7 days in blast exposed animals compared to CCI. Moreover, expression of several genes in rats subjected to noise exposure was changed compared to naive rats.
Several studies have been performed to evaluate gene expression changes in rodent brain following mechanical trauma, including CCI and fluid percussion injury (FPI), mostly cortical and hippocampal tissues [3,7-10]. Less information is available on gene pattern expression following blast in rodents. In a recent study in rats, hippocampal and prefrontal cortex changes have been assessed using a limited gene array with an emphasis on inflammatory components 24 hours after blast [11]. Risling and co-workers studied mechanisms of blast induced TBI in rats with and without head acceleration and in brain penetration injury, including genomic alterations in hippocampus at 24 hours, also with emphasis on neuroinflammation, cell death and synaptic transmission [12]. They noted however an important down-regulation of genes involved in neurogenesis and synaptic transmission. In mice, behavioral changes and accompanied expression of certain genes in hippocampus were compared after blast and mechanical trauma and then validated by Q-PCR [13].
In our study, we employed blast TBI model and compared side-by-side genomic changes in rat midbrain with CCI at 24 hours and 7 days after challenge using a full size Affimetrics GeneChip (31,100 sequences) array provided by Assuragen, Inc. Also, we analyzed significantly changed genes with ANOVA p<0.05 and grouped them into major functional clusters/ groups, (a) systemic/vascular responses; neuroinflammation; proteolysis; (b) neuromediators; signaling; ion channels; and (c) neural development/ repair; In both blast and CCI groups, t-tests were used for assessing significance at 24 h and 7 days vs respective controls and, in some cases CCI control vs blast noise control. Finally, gene interactions following blast and CCI have been mapped and depicted using Systems Biology tool. Pathways reflecting gene-associated molecular functions altered after blast injury were created and compared to CCI.
Systemic/vascular responses, neuroinflammation, proteolysis
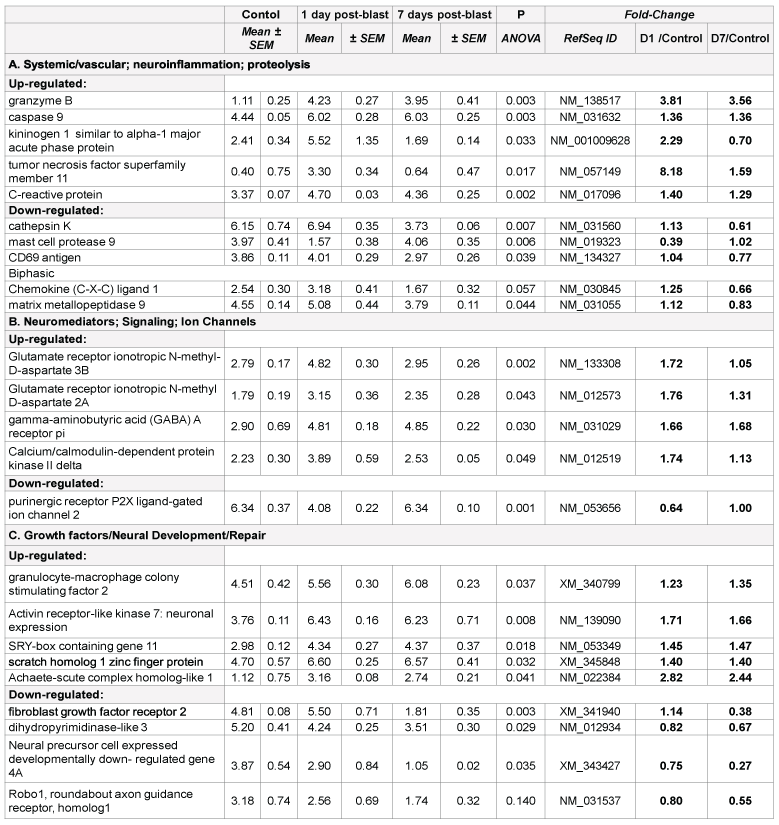
Prominent changes in several proteolysis and inflammation genes were found in both blast and CCI groups, including acute phase kininogen 1, cathepsins and chemokines (Table 1A and 2A). However, there are important disparities between blast and CCI groups in these responses. First, several chemokines were markedly up-regulated 24 h and 7 d post- CCI impact, while only C-X-C ligand 1 mRNA expression was modestly increased 24 h post-blast followed by down-regulation at day 7 after blast. Second, expression of TNF superfamily member 11 was highly increased at all times after blast exposure, but was unchanged after CCI. In contrast, MMP-9 gene was up- regulated after CCI throughout post-impact period, but slightly down-regulated at day 7 after initial modest increase at 24 h following blast (Table 1A and 2A).
There are several common regulators and pathways for both blast and CCI groups such as kininogen 1 (KNG1) linking inflammation and hemostasis, cytokines and downstream NF-kB (Figures 1 and 2). The difference is reflected by involvement of estrogen receptor 2, hemoxygenase and several chemokine network in CCI (Figure 2), while CD69, C-reactive protein and acute phase cytokines together with vasoconstrictor endothelin gene are up-regulated pathways mostly characteristic for blast exposure (Figure 1). As mentioned above, distinct directions and time-course of changes in genes encoding tumor necrosis factor (TNFSF11) and metalloproteinases (MMP-9) has been revealed.
Early up-regulation of genes encoding cytokines and chemokines (IL- 1, IL-6, C-X-C-L10) has been demonstrated in several recent studies, including gene expression profiling in hippocampus and cortex after both CCI and blast [7,11 14,15]. In addition to pro-inflammatory molecules, up-regulation of anti-inflammatory IL-10 and tissue inhibitor of metalloproteinase-1 (TIMP-1) has been shown in several reports [11,12]. In this paper, we show important differences in blast vs. CCI responses, which may have both diagnostic/prognostic significance and, more important therapeutic strategies for TBI of different genesis.

RNA samples were hybridized with fluorescent labelled probes in triplicate using Affimetrix chips and analysed with Agilient Technology scanner equipped with software (see Materials and methods for details). mRNAs were grouped in functional clusters and time-course difference of most important gene expression is presented. Background-corrected and internal control-normalized fluorescence values are shown as Mean+ SEM, of at least 3 rats in animal group, each rat sample processed in duplicate.
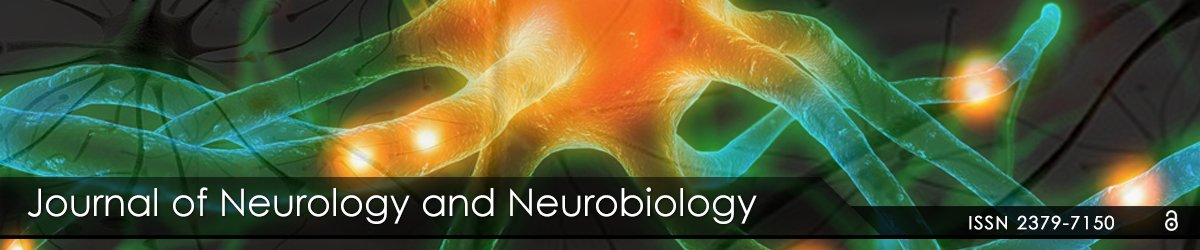
Neuromediators; Signaling; Ion Channels: Very important features of gene expression in this cluster was prominent up-regulation of S100 calcium binding protein A11 (calizzarin), S100 calcium binding protein A8 (calgranulin A), and S100 calcium-binding protein A4 after CCI, but not after blast (Table 2B). Significant up-regulation of GABA A receptor pi gene was observed in blast-exposed rats groups compared with noise control (Table 1B). There was a modest, but not significant (p=0.23) increase in this gene expression in CCI group vs. control (Table 2B). Similarly, remarkable up-regulation of glutamate ionotropic N-methyl-Daspartate receptors was found in blast-exposed groups vs control but not in CCI treated rats (Table 1B and 2B). Differential gene interactions of this cluster, including NCAM and VEGF-A, after blast, compared to S100A family after CCI is shown in Figures 1 and 2, respectively.
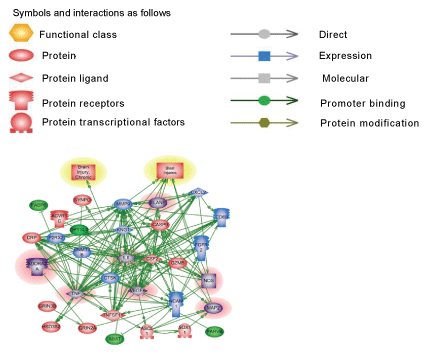
Pathways critical components that do not overlap in blast and CCI groups are shown in frames. CASP9-caspase 9; CRP- C reactive protein; IL1R2- interleukin 1 receptor type 2; ESR2-estrogen receptor type 2; HMOX1- heme oxygenase 1; CXL1-Chemokine (C-X-C) ligand 1; CXCR2- chemokine receptor. KNG1- kininogen 1; TNFSF11- tumor necrosis factor superfamily member 11. Please see text for a detailed description. The upregulated genes are shown in red and downregulated genes are in blue. Yellow highlights indicate biphasic gene regulation (see table),
Roles for S100 protein family have been demonstrated and S100B glial protein, a component of Ca2+/calmodulin signaling, has been extensively studied in TBI, including its clinical diagnostic value [16-18]. However, S100 significance has been evaluated in mostly mechanical trauma models (CCI, closed head injury, FPI) and in patients with TBI, and no attention to other components was given. Our data highlights the involvement of several S100 family genes, including calgranulin A and calizzarin pathogenesis of mechanical cortical impact, and shows a little, if any, impairment of S100 following blast exposure. Important, our data demonstrate up-regulation of glutamate ionotropic receptor genes 2A (24 h and 7 d) and 3B (24 h) over noise control, which has not been previously reported for blast exposures (Table 1B).
Growth factors/Neural Development/Repair. Significant differences between exposure groups were found in genes involved in neural development and repair. Activin receptor-like kinase 7 (neuronal expression) gene was significantly up-regulated following blast exposure vs. control group, but not in CCI-subjected group naive control. A comparable disparity is observed with Achaete-scute complex homologlike 1 and SRY-box containing gene 11 which increased after blast exposure but did not change or even down-regulated in CCI vs. naïve control (Tables 1C and 2C).
Important, expression of neural precursor cell expressed developmentally down-regulated gene 4A (Nedd4a) was significantly decreased after blast, but not changed and even slightly (but not significantly) increased after CCI compared to naïve rats. Dihydropyrimidinase-like 3 (CRMP-4) was down regulated compared respective controls in both blast and CCI groups.
The most intriguing pattern was observed in expression roundabout axon guidance receptor 1 (Robo1) gene. It was very significantly upregulated after CCI, while showing a distinct trend to down-regulate after blast exposures (Table 1C and 2C).
The pathways mapping gene interactions of this cluster in blast vs CCI are included in Figures 1 and 2 Blast exposure triggered activation of GM-CSF and downstream transcriptional activators, including SOX11 leading to induction of repair processes exemplified by tubulin, WNT (negatively) and RPS6K ribosomal protein kinase genes as well as TGFbeta related pathways, including up-regulation of ALK7 (ACVR1C)/ SMAD transcriptional regulators.
Gene-associated molecular functions
Pathways of molecular interactions in blast and CCI are shown in Figure 3. As indicated, the most important differences found in S100A family genes which were up-regulated in CCI cohort and involved in downstream regulation of ubiquitin, proteinase activated receptor, Rho, GPCR, JAK signaling. In blast-induced group, functional regulation pathways demonstrate potential NCAM1- dependent processes with glutamate receptor, CAMK and calmodulin signaling, and GABA/GABA A receptor activation (Table 1B and Figure 3). The pathology/disease maps generated using this cluster components are very similar in blast and CCI groups and overlap with cluster 1 (systemic/vascular/neuroinflammation/ proteolysis), although involve some different components- S100A for CCI and NCAM for blast (Table 2B and Figures 3 and 4).
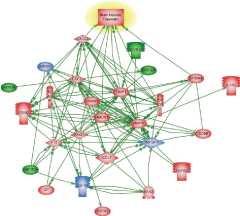
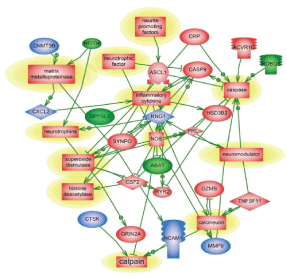
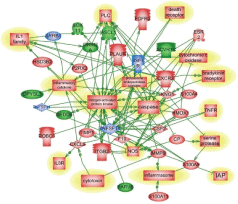
Genes and molecular processes involved in neurorepair are still unclear, thus elucidating mechanisms, molecular components and biomarkers of post-injury outcome will have immense clinical importance. FGF pathways with downstream MAP kinase family are negatively involved in gene regulation following blast exposure (Table 1C, Figure 1). After CCI, the central point of gene regulation in midbrain appears to be IGF1 and EGF family molecules, including IGF1 receptor and EGF ligand genes connecting similar to blast effector genes such as tubulin, S6 kinase, histones and WNT as shown in Table 2C and Figures 2 and 4.
The functional roles for ROBO1 and NEDD4 in CNS is not clear at present, but it appears from the map that down-regulation of ROBO1 and NEDD4 genes can facilitate PI3 kinase and RAS and stimulate repair (Figures 3 and 4), while up-regulation of ROBO1 and NEDD4 after CCI can negatively influence neural repair.
It has been shown that ROBO1 modulates neurogenesis in the developing cortex [19]. ROBO1 is involved in axonal guidance of dopaminergic neurons by neuropilin-2 [20], up-regulation of which we have shown previously [21]. However, the opposite regulation of ROBO1 after blast compared to CCI may reflect the spatial specificity in the expression and function of ROBO1 in forebrain and neocortex [22-24], which is differentially affected by blast wave in midbrain than by CCI insult directly to the cortex. However, the roles for ROBO1 and related Slit system in brain repair after TBI has not been investigated. Also, roles for Activin receptor family, including ALK kinases have not been studied following TBI, although ALK1 signaling produced lower TBI-induced increases in GFAP mRNA levels, indicating attenuation in astroglial and neuronal responses to injury [25].
ACVR1C was significantly up-regulated after blast compared to noise control, while the CCI values were not equally increased compared to naïve rats. Similarly to ROBO1, NEDD4a was down-regulated in midbrain after blast, but was up-regulated at 7 day after CCI. NEDD4 (WW) was shown to be expressed in surviving neurons after cortical injury [26], and differential expression of these genes in midbrain may reflect spatial disparities. Taken together, our data reveal for the first time that ALK7, ROBO1 and NEDD4 are involved in post-blast and post CCI, possibly brain repair. In a recent paper, Heinzelmann and co-workers reported down- regulation of several growth factors and receptors genes, such as EGFR recepror and tensin-1 in peripheral blood collected from patients with post-blast syndrome [27]. Specific mechanisms involving these important components in neural injury and recovery after TBI remain to be investigated.
Conclusion
To the best of knowledge, this is the first genomics study that assesses altered gene expression side-by-side in 2 well-defined and characterized brain insults- the TBI modeled via blast overpressure vs the CCI open head injury. For this purpose, a comprehensive study utilizing systems biologybased analysis of transcriptomics in rat midbrain has been conducted to compare blast exposure and mechanical cortical impact brain insult. Our data show both an overlap and significant time-dependent differences in gene expression changes among these groups, particularly in genes of neural development and repair. Novel components, such as ROBO1 and NEDD4 have been shown to be expressed in opposing manner following TBI induced by blast compared to CCI, suggesting different mechanisms may be involved in neurorepair dynamics. The results obtained suggest further in-depth studies are required to elucidate differences in mechanisms of brain injury produced by blast compared to mechanical impact. This will help to reveal potential biomarkers and develop diagnostics of chronic posttraumatic encephalopathy.
Acknowledgements
The authors wish to thank Olena Glushakova for excellent technical assistance. This work was supported by grants W81XWH-8-1-0376 and W81XWH-07-01-0701 from Department of Defence. The research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. K.W.W. and S.I.S are shareholders of Banyan Biomarkers, Inc., a company interested in commercializing diagnostic tests for traumatic brain injury.
Required disclaimer:
The views and opinions expressed in this manuscript are those of the author(s) and do not reflect official policy or position of the Department of Defence or the U.S. Government.
References
- Bramlett HM, Dietrich WD (2014) Long-Term Consequences of Traumatic Brain Injury: Current Status of Potential Mechanisms of Injury and Neurologic Outcomes. J Neurotrauma.[Ref.]
- Barr TL, Alexander S, Conley Y (20110) Gene expression profiling for discovery of novel targets in human traumatic brain injury. Biol Res Nurs 13: 140-153. [Ref.]
- Di Pietro V, Amin D, Pernagallo S, Lazzarino G, Tavazzi B, et al. (2010) Transcriptomics of traumatic brain injury: gene expression and molecular pathways of different grades of insult in a rat organotypic hippocampal culture model. J Neurotrauma 27: 349-359.[Ref.]
- Redell JB, Moore AN, Grill RJ, Johnson D, Zhao J, et al. (2013) Analysis of functional pathways altered after mild traumatic brain injury. J Neurotrauma 30: 752-764.[Ref.]
- Svetlov SI, Prima V, Kirk DR, Gutierrez H, Curley KC, et al. (2010) Morphologic and biochemical characterization of brain injury in a model of controlled blast overpressure exposure. J Trauma 69: 795-804.[Ref.]
- Glushakova OY, Johnson D, Hayes RL (2014) Delayed increases in microvascular pathology after experimental traumatic brain injury are associated with prolonged inflammation, blood-brain barrier disruption, and progressive white matter damage. J Neurotrauma 31: 1180-1193.[Ref.]
- Almeida-Suhett CP, Li Z, Marini AM, Braga MF, Eiden LE (2014) Temporal course of changes in gene expression suggests a cytokine related mechanism for long-term hippocampal alteration after controlled cortical impact. J Neurotrauma 31: 683-90.[Ref.]
- Anderson GD, Farin FM, Bammler TK, Beyer RP, Swan AA (2011) The effect of progesterone dose on gene expression after traumatic brain injury. J Neurotrauma 28: 1827-43.[Ref.]
- Colak T, Cine N, Bamac B, Kurtas O, Ozbek A, et al. (2012) Microarraybased gene expression analysis of an animal model for closed head injury. Injury 43: 1264-1270.[Ref.]
- White TE , Ford GD, Surles-Zeigler MC, Gates AS, LaPlaca MC, et al. (2013) Gene expression patterns following unilateral traumatic brain injury reveals a local pro-inflammatory and remote anti-inflammatory response. BMC Genomics 14: 282.[Ref.]
- Kochanek PM, Dixon CE, Shellington DK, Shin SS, Bayır H, et al. (2013) Screening of biochemical and molecular mechanisms of secondary injury and repair in the brain after experimental blastinduced traumatic brain injury in rats. J Neurotrauma 30: 920-937.[Ref.]
- Risling M, Plantman S, Angeria M, Rostami E, Bellander BM, et al. (2011) Mechanisms of blast induced brain injuries, experimental studies in rats. Neuroimage 1: S89-97.[Ref.]
- Tweedie D, Rachmany L, Rubovitch V, Zhang Y, Becker KG, et al. (2013) Changes in mouse cognition and hippocampal gene expression observed in a mild physical- and blast-traumatic brain injury. Neurobiol Dis 54: 1-11.[Ref.]
- Kane MJ, Angoa-Pérez M, Francescutti DM, Sykes CE, Briggs DI, et al. (2012) Altered gene expression in cultured microglia in response to simulated blast overpressure: possible role of pulse duration. Neurosci Lett 522: 47-51.[Ref.]
- Valiyaveettil M, Alamneh YA, Miller SA, Hammamieh R, Arun P, et al. (2013) Modulation of cholinergic pathways and inflammatory mediators in blast- induced traumatic brain injury. Chem Biol Interact 203: 371-375.[Ref.]
- Donato R, Cannon BR, Sorci G, Riuzzi F, Hsu K, et al. (2013) Functions of S100 proteins. Curr Mol Med 13: 24-57.[Ref.]
- Papa L, Ramia MM, Kelly JM, Burks SS, Pawlowicz A, et al. (2013) Systematic review of clinical research on biomarkers for pediatric traumatic brain injury. J Neurotrauma 30: 324-338.[Ref.]
- Serpero LD, Bellissima V, Colivicchi M, Sabatini M, Frigiola A, et al. (2013) Next generation biomarkers for brain injury. J Matern Fetal Neonatal Med 2: 44-49.[Ref.]
- Yeh ML, Gonda Y, Mommersteeg MTM, Barber M, Ypsilanti AR, et al. (2014) Robo1 modulates proliferation and neurogenesis in the developing neocortex. J Neurosci 34: 5717-5731.[Ref.]
- Torigoe M, Yamauchi K, Tamada A, Matsuda I, Aiba A, et al. (2013) Role of neuropilin-2 in the ipsilateral growth of midbrain dopaminergic axons. Eur J Neurosci 37: 1573-1583.[Ref.]
- Svetlov SI, Prima V, Glushakova O, Svetlov A, Kirk DR, et al, 2012 Neuro-glial and systemic mechanisms of pathological responses in rat models of primary blast overpressure compared to “composite” blast. Front Neurol 3: 15.[Ref.]
- Alpár A, Tortoriello G, Calvigioni D, Niphakis MJ, Milenkovic I, et al. (2014) Endocannabinoids modulate cortical development by configuring Slit2/Robo1 signalling. Nat Commun 5: 4421.[Ref.]
- Borrell V, Cárdenas A, Ciceri G, Galcerán J, Flames N, et al. (2012) Slit/Robo signaling modulates the proliferation of central nervous system progenitors. Neuron 76: 338-352.[Ref.]
- Hernández-Miranda LR1, Cariboni A, Faux C, Ruhrberg C, Cho JH, et al. (2011) Robo1 regulates semaphorin signaling to guide the migration of cortical interneurons through the ventral forebrain. J Neurosci 31: 6174-6187.[Ref.]
- Israelsson C, Lewén A, Kylberg A, Usoskin D, Althini S, et al. (2006) Genetically modified bone morphogenetic protein signalling alters traumatic brain injury-induced gene expression responses in the adult mouse. J Neurosci Res 84: 47-57.[Ref.]
- Sang Q, Kim MH, Kumar S, Bye N, Morganti-Kossman MC, et al. (2006) Nedd4-WW domain-binding protein 5 (Ndfip1) is associated with neuronal survival after acute cortical brain injury. J Neurosci 26: 7234-7244.[Ref.]
- Heinzelmann M, Reddy SY, French LM, Wang D, Lee H, et al. (2014) Military personnel with chronic symptoms following blast traumatic brain injury have differential expression of neuronal recovery and epidermal growth factor receptor genes. Front Neurol 5: 198.[Ref.]
- Agoston DV, Elsayed M (2012) Serum-based protein biomarkers in blast-induced traumatic brain injury spectrum disorder. Front Neurol 3: 107.[Ref.]
- Ou S, Liu GD, Zhou LS, Xia X, Bai SR, et al. (2014) Bioinformatics analysis of gene expression profiles in the rat cerebral cortex following traumatic brain injury. Eur Rev Med Pharmacol Sci 18: 101-107.[Ref.]
- Walder B, Robin X, Rebetez MM, Copin JC, Gasche Y, et al. (2013) The prognostic significance of the serum biomarker heart-fatty acidic binding protein in comparison with s100b in severe traumatic brain injury. J Neurotrauma 30: 1631-1637.[Ref.]
Download Provisional pdf here
SCI FORSCHEN JOURNALS
All Sci Forschen Journals are Open Access
