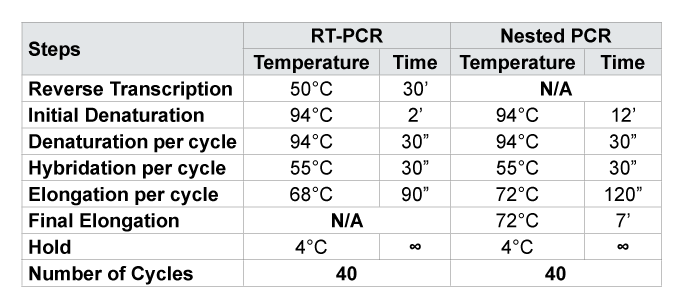
N/ANot Applicable for this amplification; 30’ = 30 minutes; 30” = 30 seconds

Erick Ntambwe Kamangu,1* Adawaye Chatté,2 Fabrice Susin,3 Raphael Boreux,4 Richard Lunganza Kalala,1 Georges Lelo Mvumbi,1 Patrick De Mol,4 Dolores Vaira,3 Marie-Pierre Hayette,3,4
1Molecular Biology Unit, Department of Basic Sciences, Faculty of Medicine, University of Kinshasa (UNIKIN), Kinshasa, Democratic Republic of Congo (DRC)*Corresponding author: Erick Ntambwe Kamangu, Molecular Biology Unit, Department of Basic Sciences, Faculty of Medicine, University of Kinshasa (UNIKIN), Kinshasa, Democratic Republic of Congo (DRC); E-mail: erick.kamangu@unikin.ac.cd or erickamangu@gmail.com
Background
The massive use of antiretroviral (ARV) created the emergence of mutant strains resistant to treatment. Thus, the World Health Organization (WHO) recommends epidemiological monitoring for newly infected patients with the Human Immunodeficiency Virus (HIV). The objective of this study is to determine the genetic diversity of HIV Type 1 and the prevalence of mutations associated with resistance to ARV in treatment-naïve patients in Kinshasa. The massive use of antiretroviral (ARV) created the emergence of mutant strains resistant to treatment. Thus, the World Health Organization (WHO) recommends epidemiological monitoring for newly infected patients with the Human Immunodeficiency Virus (HIV). The objective of this study is to determine the genetic diversity of HIV Type 1 and the prevalence of mutations associated with resistance to ARV in treatment-naïve patients in Kinshasa.Methods
One hundred fifty-three subjects diagnosed positive for HIV Type 1 voluntarily participated in this study. They were recruited in different centers in Kinshasa. The inclusions were conducted from August 2013 to February 2014. Five milliliters (5 ml) of blood were collected in a tube with anticoagulant EDTA. Five hundred microliter of plasma was sent for analysis to the AIDS Reference Laboratory of the University Hospital of Liège (CHU-Liège) in Belgium. RNA was extracted from 140 µl of plasma using the QIAamp RNA Mini Kit (QIAGEN®). A Reverse Transcriptase PCR and Nested PCR enabled amplification of the regions of interest on the Protease and Reverse Transcriptase (RT) for subsequent sequencing.Results
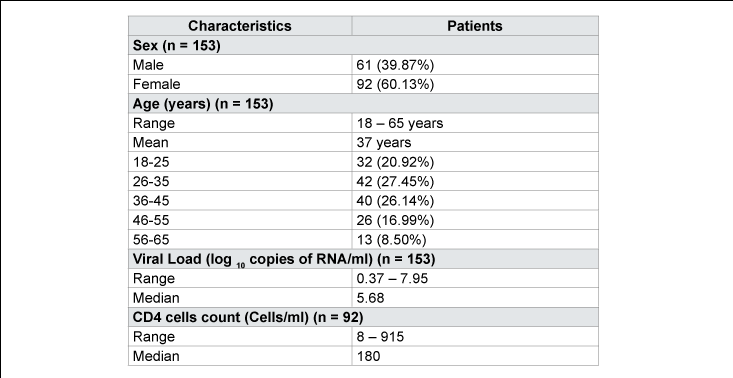
The mean age of patients was 37 years, ranging from 18 to 65 years. The median values of Viral Loads (VL) and rate of CD4 lymphocytes were respectively 5.68 log10 RNA copies/ml and 180 cells/ml. Protease and RT were amplified and sequenced, respectively, for 130 (84.9%) and 145 (94.8%) patients out of 153. Subtype A was dominant with 35 cases (22.9%); followed by CRF02_AG (11.1%), C (9.8%), G (9.8%), K (9.8%), D (7.8%), H (7.8%) and J (5.0%).Conclusion
The results of our study confirm the high diversity of HIV Type 1 in Kinshasa. It reveals the heterogeneity and the dynamic of the virus, and the presence of transmitted resistance associated with antiretroviral drugs.HIV-1; Genetic Diversity; ARV Resistance; Naïve patients; Kinshasa
The Human Immunodeficiency Virus (HIV) is classified according to the types of the viruses, groups, subtypes, sub-subtypes and Circulating Recombinant Forms (CRFs) [1-3]. This classification has the advantage of better choose the type of treatment appropriate to the infected patient [2,3]. In Sub-Saharan Africa, molecular epidemic is very diverse and heterogeneous; it is dominated by the subtypes A, C, G and different CRFs [1-6].
The introduction of Antiretroviral drugs (ARVs) reduced the morbidity and mortality associated with HIV infection [7]. But also, its massive and disproportionate use caused the emergence of mutant strains resistant to treatment. Thus, the World Health Organization (WHO) recommended epidemiological monitoring for newly infected patients with HIV [7]. This includes monitoring the prevalence of different circulating strains and the search for mutations associated with treatment in the population.
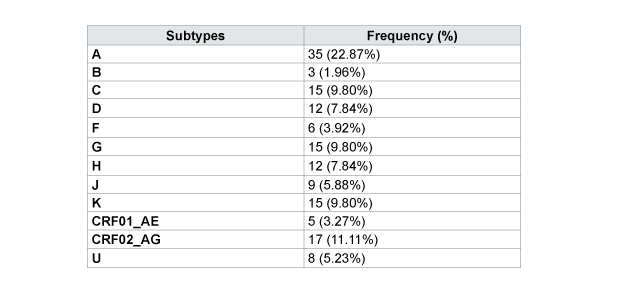
Kinshasa is a city with moderately low HIV prevalence; it has been below 5% for the 5 past years [8]. Introduced in 2002, access to ARVs is still near to the ground, less than 15% of the eligible population has access to the treatment [8]. In the Democratic Republic of Congo (DRC), the recommended therapy is a combination of 2 Nucleotide Reverse Transcriptase Inhibitors (NRTI) and 1 Non-Nucleotide Reverse Transcriptase Inhibitor (NNRTI) for the first-line treatment [9]. For the second-line treatment, it is of 1 Protease Inhibitor (PI) and 2 NRTI [9]. The most commonly used molecules for NRTI are: Abacavir (ABC), Didanosine (ddI), Lamivudine (3TC), Stavudine (d4T) and Zidovudine (ZDV); for NNRTI: Efavirenz (EFV) and Nevirapine (NVP); and for PI: Lopinavir boosted by Ritonavir (LPV/r) [10]. LPV/r is introduced only in the second intension treatment [9,10]. In Kinshasa, the variants A (34.3%) dominatedthe epidemiology in the literature; they are followed by variants G (17.3%), D (11.2%), C (7.4%), CRF01_AE (4.9%), F (4.8%), H (3.9 %) and U (3.5%) [11].
The objective of this study is to determine the genetic diversity of HIV Type 1 and the prevalence of mutations associated with resistance to antiretrovirals (ARVs) in treatment-naïve patients in Kinshasa.
Population
One hundred and fifty-three (153) subjects diagnosed positive for HIV Type 1 voluntarily participated in this study. They were recruited in different centers of Kinshasa [10]. The inclusion criteria for subjects were: (i) be diagnosed HIV-1 positive according to national guidelines [9], (ii) be over 18 years old at the inclusion, (iii) be eligible for the Antiretroviral therapy (ART) in the monitoring center and (iv) be naive to ART according to the records of the centers. Nested PCR DNA was used to confirm HIV positivity for the patients in the laboratory of Molecular Biology at the University of Kinshasa [12]. The patient’s demographic, clinical and laboratory information were recorded on standardized sheets pre-tested for the study. All patients signed an informed consent form for participation in the study. The inclusions were conducted from August 2013 to February 2014.
Blood samples
Five milliliters (5 ml) of blood were collected in a tube with anticoagulant EDTA from the vein of the elbow crease. The collected blood was centrifuged at 1000 g for 10 minutes to obtain a clear separation in 3 phases (plasma, buffy coat and Cullot). One milliliter of plasma (supernatant) was transferred into a microtube previously labeled and sent to the laboratory of Molecular Biology of the Faculty of Medicine for analysis. Samples were aliquoted into 2 tubes of 500μl each. One tube was used to determine the Viral Load (VL) of patients at baseline in Kinshasa [13]. The other tube was sent to the AIDS Reference Laboratory of the University Hospital of the University of Liège (CHU-ULg) in Liège, Belgium for sequencing.
CD4 count was done with the Facs Count (Becton Dickinson Immunocytometry Systems) at the respective centers for patients.
RNA extraction, amplification and sequencing
RNA was extracted from plasma 140 µl using the QIAamp RNA Mini Kit (QIAGEN® ) at the LRS CHU-Ulg [14]. The extracted samples were stored at -80°C until use.
After extraction, a Reverse Transcription PCR (RT-PCR) and Nested PCR were performed to amplify the regions of interest on the Protease and Reverse Transcriptase (RT) for subsequent sequencing. PCR were performed under the conditions of cycles and temperatures previously described by Steegen (Table 1) [15]. Each sample was amplified on the Protease and Reverse Transcriptase in the sense and antisense. The primers used for PCR assays are presented in Table 2. For the samples which were not amplified with the first primers (Normals), other PCR were done with Alternatives primers (ATL) previously described in the literature [15].
The amplified fragments obtained are approximately 500 base pairs for the protease and 800 base pairs for the RT. These fragments were sequenced by the Sanger sequencing method. They were purified by the ExoSAP-IT technique to eliminate the residual products of the PCR amplification and sequenced by the purified fragment. A second purification of the product was done before the reading of the sequence.
The pairing of the resulting fragments (sense and antisense) was performed with the software Vector NTI Advance® 11.5 (Invitrogen, Life Technologies) and compared with the Stanford University database (hivdb.stanford.edu) [16], that of the French National Agency for Scientific Research (ANRS - www.hivfrenchresistance.org) and that of the Max Plank (www.geno2pheno.org) for identification of the subtypes of HIV-1 and possible mutations. Minor adjustments of the bases were performed if necessary to align the sequences.
Statistics
The Friedman test was used to analyze differences in the groups variants and different mutations. The probability p value <0.05 was considered significant. Viral load is expressed in decimal logarithm per ml.
One hundred and fifty three (153) patients infected with HIV Type 1 were selected for this study. Our population is composed of 61 (39.9%) men and 92 (60.1%) women; is a sex ratio of 1.5 women per man (p<0.05). The average age was 37 years, ranging from 18 and 65 (Table 3). The median values of Viral Loads (VL) and rate of CD4 lymphocytes were respectively 5.68 log10 RNA copies/ml and 180 cells/ml (Table 3).
Subtyping
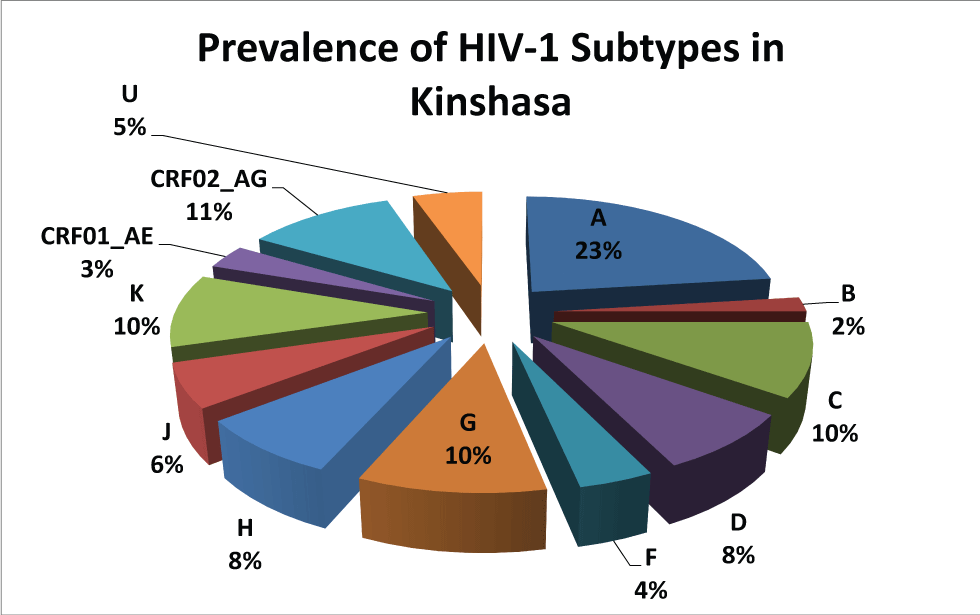
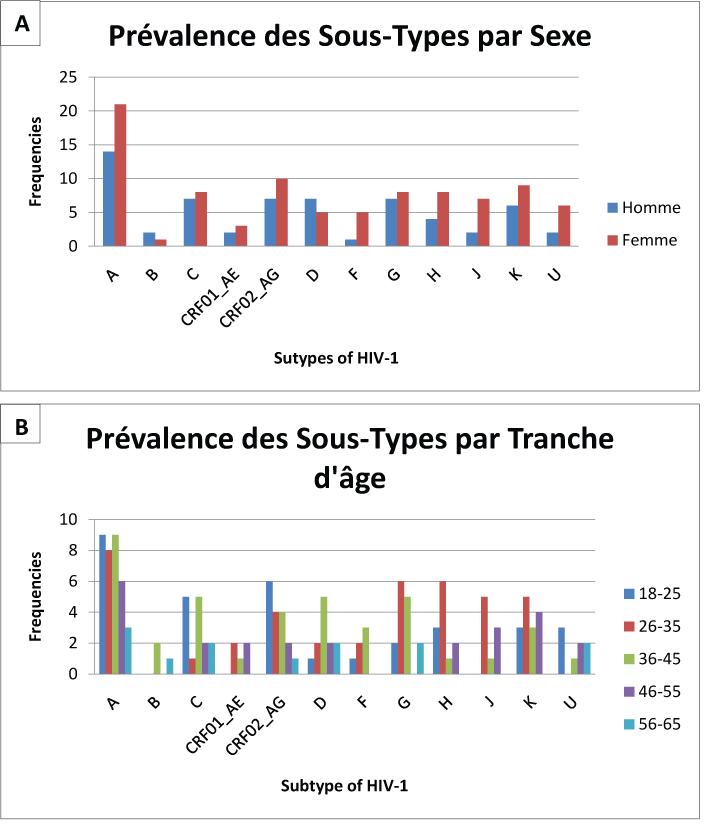
Protease and Reverse Transcriptase were amplified and sequenced, respectively, for 130 (84.9%) and 145 (94.8%) patients. Hundred twenty three (123) samples were amplified with the normal primers and 7 with alternatives to the region of the protease, respectively. Hundred and twenty five (125) samples were amplified with normal primers and 20 with alternatives for the region of RT, respectively. Subtype A is dominant with 35 cases (22.9%); followed by CRF02_AG (11.1%), C (9.8%) G (9.8%) K (9.8%), D (7.8%), H (7.8%) and J (5.0%) as described in Table 4 and Figure 1. There is not a large difference in the distribution of subtype by sex; it is mostly dominated by female except for the subtype B and D (Figure 2). The distribution of subtype by age interval is presented in Figure 2.
Mutations associated with protease inhibitors (PIs)
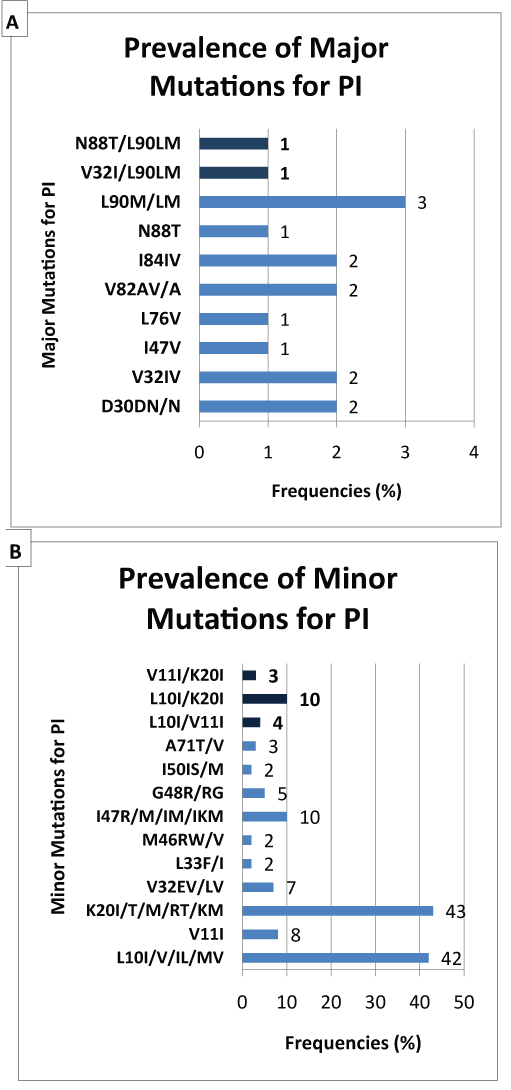
Mutations associated with resistance to protease inhibitors can be classified into majors or minors. In our study, we observed several mutations associated with protease inhibitors (Figure 3). The most significant major mutations are: L90M (2.0%), D30N (1.3%), V32I (1.3%), V82A (1.3%) and I84V (1.3%). The most common minor mutations are: K20I/RT/M (28.1%), L10I (27.5%), I47R/IM (6.5%), V11I (5.2%), V32E/L (4.6%), G48R (3.3%) and A71T/V (2.0%).

N/ANot Applicable for this amplification; 30’ = 30 minutes; 30” = 30 seconds
Associated mutations Reverse Transcriptase Inhibitors (RTI)
We also observed mutations associated with Nucleoside Reverse Transcriptase Inhibitors (NRTI) and Non-Nucleotide Reverse Transcriptase Inhibitors (NNRTI) (Figure 4). The most frequent mutations for NRTI are: V75I/D/L/M (18.3%), K70E/N/R (9.8%), D67G/ E/N (9.2%), M184V/L/K/R (9.2%), T215F/N/I/L (9.2%), Y115F (7.8%), M41L (7.2%), T69P/N (5.2%) and L74V/M/I (3.9%). The most recurrent mutations for NNRTI are: V179F/T/D (9.8%), K103N/I/NRS (8.5%), V106I/A (7.2%), Y181K/C (5, 8%), V90I/GIS (5.8%), A98G/GPR (5.2%), V108I/KN (5.2%), Y188C/L/D (4.6%) and F227C/L (4.6%).
The aim of this study was to determine the genetic diversity of HIV Type 1 and the prevalence of mutations associated with resistance to Antiretroviral drugs (ARVs) in treatment-naïve patients in Kinshasa. Our population consisted of 61 (39.9%) men and 92 (60.1%) women; a sex ratio of 1.5 women per man (p<&0.05). The age group most represented age is that of 26 to 35 years with 42 patients (27.45%), followed by 36 to 45 years with 40 patients (26.14%). The same socio-demographic information about the infected population are found in different studies in our environment [8,9,17,18].
Out of 153 patient’s naïve of Antiretroviral Treatment (ART), we amplified 130 samples (84.9%) on the region of the protease and 145 samples (94.8%) in the region of the Reverse Transcriptase (RT). This gives us an amplification rate higher than 85% for the two regions. This difference in amplification was also observed in various conditions; samples are more easily amplified on the Reverse Transcriptase than the protease because of the size of the gene of interest [19,20]. Amplification failures can be due to the high diversity of the amplification site of HIV type 1 variants that exist in Kinshasa [11]. They can also be caused by low Viral loads (VL) or under the detection limit [15,19]. Indeed, all 7 samples (4.6%) who had a VL less than 3.0 log10 RNA copies/ml did not give any amplification.
Subtype A is dominant in Kinshasa, up to 22.9% of the study population. It is followed by CRF02_AG (11.1%), C (9.8%) G (9.8%) K (9.8%), D (7.8%), H (7.8%) and J (5.0%) as described in Table 4. Our results are consistent with the different subtypes found in the Central African countries [4,11,21] and in the neighboring countries of Western Africa [4,11,20-22]. Apart from the high prevalence of CRF02_AG, our results are consistent with the literature reported in the past for Kinshasa [11,22- 24]. The strong presence of the recombinant form CRF02_AG reflects the dynamism of HIV in Kinshasa. This can originate from rural population movement to the metropolitan city, but also from the return of expatriates and immigrants in the capital city; thus bringing more diversity in the city. The distribution of the subtypes by sex and by age presents an interesting mosaic. Subtype B is mostly found in male and in the age interval of 36 to 45 years and that of 56 to 65 years. Most of the others are presented according to the sex-ratio and the age. There is no significant association between HIV-1 subtypes and age or sex in our population of study.
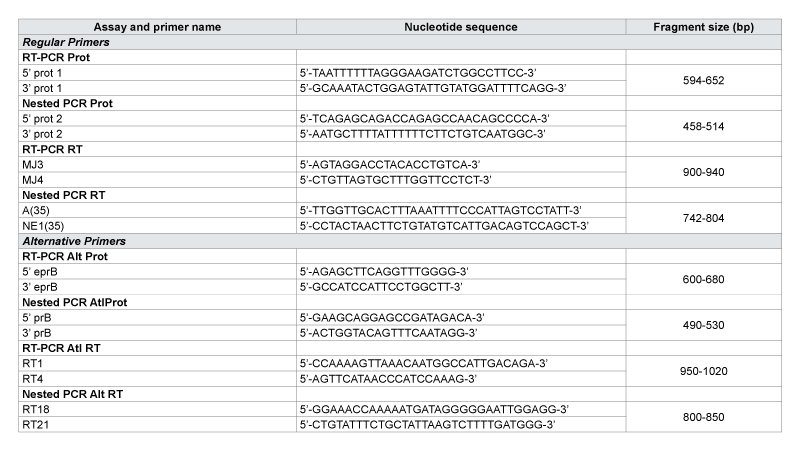
Prot: Protease; RT: Reverse Transcriptase; Alt: Alternative primers
Several primary resistance were observed for PI; major and minor mutations. Genetic barrier is high for this class of drugs; one major mutation for PI can lead to irreversible resistance for the specific PI. While it takes an accumulation of several minor mutations to lead to irreversible resistance for PI. The most major mutations observed are: L90M (2.0%), D30N (1.3%), V32I (1.3%), V82A (1.3%) and I84V (1.3%). The most minor mutations are: K20I/RT/M (28.1%), L10I (27.5%), I47R/M (6.5%), V11I (5.2%), V32E/L (4.6%), G48R (3.3%) and A71T/V (2.0%). PIs are used in the DRC only for the second line as recommended by the national program [9]. These high frequencies of minor mutations transmitted ART-naïve patients are alarming especially since they can impart to patients resistant to treatment in second intension in the case of regime change. If the first line of treatment failure, almost 2.0% of patients will develop an irreversible resistance to second-line ART regimen that includes Lopinavir boosted by Ritonavir (LPV/r); implying a failure to this line of treatment too.
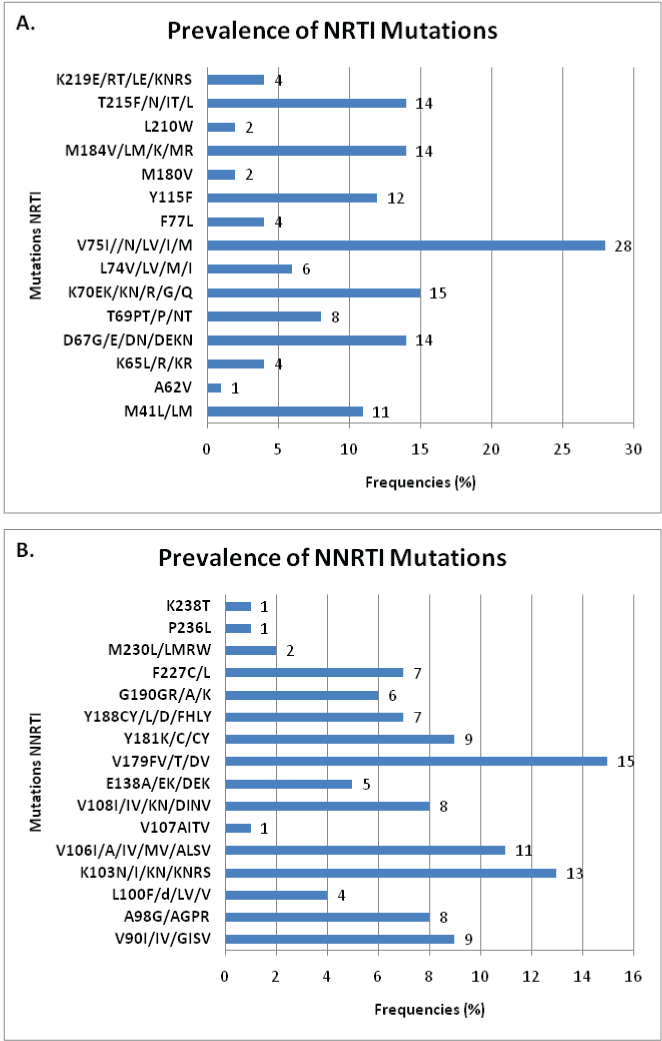
K65R mutation (2.6%) is associated with resistance to Stavudine (d4T), Abacavir (ABC), Lamivudine (3TC), Tenofovir (TDF) and Didanosine (ddI); the K70E/N/R (9.8%) is associated with the TDF; the L74V/M/I (3.9%) with ddI and ABC; the V75I/D/L/M (18.3%) with d4T; the Y115F (7.8%) with ABC; the M184V/L/K/R (9.2%) with 3TC; and T215F/N/I/L (9.2%) with Zidovudine (ZDV) and d4T [25-36]. For NNRTI, mutation A98G (5.2%) is associated with resistance to Nevirapine (NVP) and the E138A/K (3.3%) is documented for the resistance to Efavirenz (EFV); the L100F/V (2.6%), the K103N (8.5%), the V106I/A/M (7.2%), the Y181K/C (5.8%), the Y188C/L/D (4.6%), the G190R/A (3.9%) and M230L (1.3%) are all associated with resistance to both NVP and EFV [28,32,35,37- 40]. We noticed that at this point, nearly 10% of naive patients have a predisposition to first-line treatment failure as recommended for the DRC (ZDV + 3TC + NVP) due to transmitted mutations.
The most incriminated mutations for resistance to first-line ART in DRC have varying prevalence: V75I (18.3%), K70E (9.8%), D67N (9.2%), M184V (9.2 %), T215F (9.2%), Y115F (7.8%), M41L (7.2%), T69P (5.2%) and L74VI (3.9%) for the NRTI; V179F (9.8%), K103N (8.5%), V106A (7.2%), Y181C (5.8%), V90I (5.8%), A98G (5.2%), V108I ( 5.2%), Y188C (4.6%) and F227C (4.6%) for NNRTI; and: L90M (2.0%), D30N (1.3%), V32I (1.3%), V82A (1.3%) and I84V (1.3%) for PI.
The results of our study show a strong diversity of HIV Type 1 in Kinshasa. It reveals heterogeneity and dynamic of the virus, and presence of primary resistance associated with antiretroviral drugs. Several major and minor resistances associated with protease inhibitors, as well as mutations associated with reverse transcriptase inhibitors were detected in patient’s naïve of antiretroviral therapy. This demonstrates that genotyping tests must be made before starting patients on treatment and before the decision to change the treatment for better care for patients.
Conflict of interest
The authors declare that they have no conflicting interest.
Authors’ contribution
EK, AC and DV conceived the study. MPH, RK, PDM and GM participated in the study design. EK coordinated data collection. EK, AC and FS performed laboratory work, data analysis and interpretation of results. FS and RB participated in laboratory analysis. DV coordinated laboratory analysis. FS, RB, AC and DV participated in interpretation of results. EK drafted the manuscript. All authors contributed to the writing, read and approved the final manuscript.
Download Provisional PDF Here
Article Type: Research Article
Citation: Kamangu EN, Chatte A, Susin F, Boreux R, Kalala RL, et al. (2015) Genetic Diversity and Antiretroviral Drug Resistance among Drug-Naïve HIV Type 1 Infected Patients attending Clinics in Kinshasa, Democratic Republic of Congo. J HIV AIDS Volume1.1: http://dx.doi.org/10.16966/2380-5536.101
Copyright: © 2015 Kamangu EN et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Publication history:
All Sci Forschen Journals are Open Access