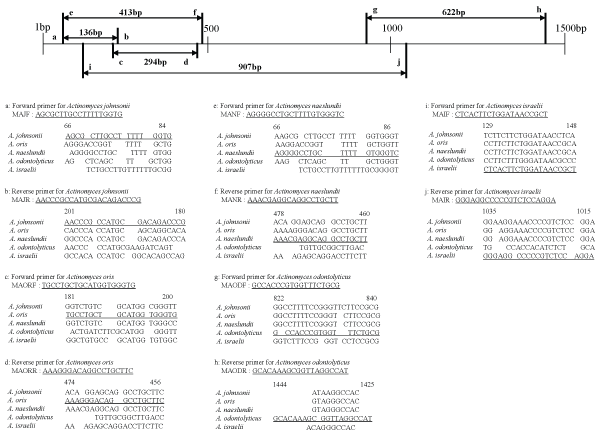
Figure 1: Locations and sequences of species-specific primers for the 16S rDNA of group A oral Actinomyces species. The nucleotide sequence of each primer has been underlined.


Taira Kobayashi1* Satoshi Uchibori1 Osamu Tsuzukibashi2 Chihiro Uezato1 Haruhiko Goto1 Chiho Mashimo3 Takayuki Nambu3 Koji Umezawa4 Mitsuhiro Ohta5
1Department of Crown Bridge Prosthodontics, Nihon University, School of Dentistry at Matsudo, Chiba, Japan*Corresponding author: Taira Kobayashi, Department of Crown Bridge Prosthodontics, Nihon University, School of Dentistry at Matsudo, Chiba, Japan, Tel: +81-47-360-9383; E-mail: kobayashi.taira@nihon-u.ac.jp
Background: Actinomyces is one of the predominant genera in the oral cavity. Oral Actinomyces species play a central role in the initial stages of biofilm formation on teeth; however, limited information is currently available on the distribution of individual species in different sites or clinical conditions. Moreover, a suitable method has yet to be developed to identify oral Actinomyces species because of the phenotypic and genetic similarities between these microorganisms.
Objective: Actinomyces naeslundii, A. odontolyticus, A. oris, A. georgiae, A. gerencseriae, A. graevenitzii, A. dentalis, A. johnsonii, A. israelii, and A. meyeri among the genus Actinomyces are regarded as normal human oral Actinomyces species. The purpose of the present study was to design primers to identify oral Actinomyces species using multiplex PCR.
Methods: Polymerase chain reaction (PCR) primers were designed based on partial sequences of the 16S rDNA genes of the representative oral Actinomyces species. The 16S rDNA genes of the representative oral Actinomyces species were obtained from the DNA Data Bank of Japan, and a multiple sequence alignment analysis was performed with the CLUSTAL W program. Homology among the primers selected for oral Actinomyces species and their respective 16S rRNA sequences was confirmed by a BLAST search.
Results: These primers were able to distinguish each oral Actinomyces species and did not display cross-reactivity with representative oral bacteria or other Actinomyces species. Moreover, we developed a multiplex PCR method with the ability to identify and differentiate oral Actinomyces species (i.e., A. naeslundii, A. johnsonii, A. oris, A. odontolyticus, A. israelii, A. georgiae, A. dentalis, A. graevenitzii, A. gerencseriae, and A. meyeri) using only two PCR tubes per sample.
Conclusion: The present results indicate that our multiplex PCR method with these primers is useful for identifying the representative oral Actinomyces species. This method is easy because the use of Mighty Amp DNA Polymerase Ver.2 (Takara) means that DNA extraction may be avoided, and species identification using this method only takes approximately 2 hours. Thus, the method described herein will allow the prevalence of oral Actinomyces species and their involvement in oral infections to be fully clarified in future studies.
Genus Actinomyces; Multiplex PCR; Oral cavity; 16S rDNA
Although the genus Actinomyces was already described in 1919, many new species have recently been discovered. The genus Actinomyces currently comprises 47 species and 2 subspecies (http://www.bacterio.net/actinomyces.html). Actinomyces consist of Gram-positive, anaerobic, and aerotolerant, non-spore-forming, non-motile pleomorphic rods with various degrees of branching. Actinomyces species are frequently found as members of the normal micro-flora, particularly in the oral cavity of humans; however, they are also etiological agents in infections, such as in classical actinomycosis, human bite wounds and abscesses at different body sites, eye infections, and oral, genital, and urinary tract infections [1,2]. The detection of these microorganisms in clinical specimens is important because it may affect the prognosis and management of patients; however, difficulties are associated with identification by conventional biochemical methods.
Ten species (i.e., Actinomyces naeslundii, A. odontolyticus, A. oris, A. georgiae, A. gerencseriae, A. graevenitzii, A. dentalis, A. johnsonii, A. israelii, and A. meyeri) among the genus Actinomyces are regarded as normal human oral bacteria [3]. Their distribution in different oral sites and their role in common oral afflictions, particularly periodontal diseases, remains controversial. Previous studies successfully induced destructive periodontitis in gnotobiotic rodents with A. naeslundii [4], suggesting a potential role for these organisms in periodontal diseases. However, an elevated level of these microorganisms in inactive lesions has also been reported [5]. Recent studies revealed that Actinomyces species are highly prevalent in supra- and sub-gingival plaques in adult periodontitis [6] and gingivitis [7]. Therefore, the data available on the role of Actinomyces species in periodontal diseases are equivocal. In addition to periodontal diseases, these microorganisms have been implicated in the pathogenesis of root surface caries and refractory periapical actinomycosis [8]. Therefore, the precise detection and speciation of these microorganisms is a prerequisite for the clarification of their pathogenic potential and possibly therapeutic interventions for the aforementioned disease entities.
The accurate identification and enumeration of Actinomyces species are required in order to clarify their role in oral ecology and dental diseases. Although conventional biochemical assays are used to identify Actinomyces species, they are often imprecise due to the phenotypic variations displayed by these bacteria. Although a sequence analysis of several target genes is the most reliable method, it is expensive, laborious, and time-consuming. Thus, a simple and more reliable assay for identifying oral Actinomyces species is desired. The purpose of the present study was to design primers for the identification of oral Actinomyces species using multiplex PCR.
The following bacterial strains were used in the present study: A. naeslundii ATCC 12104, A. oris ATCC 27044, A. johnsonii JCM 16129, A. odontolyticus ATCC 17929, A. israelii ATCC 12102, A. georgiae DSM 6843, A. dentalis DSM 19115, A. graevenitzii DSM 15540, A. gerencseriae JCM 12963, A. meyeri ATCC 35568, A. viscosus ATCC 15987, Streptococcus oralis ATCC 10557, S. salivarius JCM 5707, S. anginosus ATCC 11391, S. mutans NCTC 10449, S. sobrinus ATCC 33478, Rothia dentocariosa JCM 3067, Rothia mucilaginosa JCM 10910, Corynebacterium matruchotii ATCC 14266, C. durum ATCC 33449, Neisseria sicca ATCC 29256, Aggregatibacter actinomycetemcomitans ATCC 33384, Staphylococcus aureus JCM 2874, and S. epidermidis ATCC 2414. These strains were maintained by cultivating them in brain heart infusion agar (BHI; Difco Laboratories, Detroit, Mich.) supplemented with 1% yeast extract (BHIY). Bacteria were cultured in BHIY broth at 37 °C for 24 h under anaerobic conditions (85% N2 , 10% H2 , 5% CO2 ) for members of the genus Actinomyces and in a 5% CO2 atmosphere for other representative oral bacteria.
In the present study, ten oral Actinomyces species were divided into two groups for a multiplex PCR method. Group A consisted of A. naeslundii, A. johnsonii, A. oris, A. odontolyticus, and A. israelii, and group B consisted of A. georgiae, A. dentalis, A. graevenitzii, A. gerencseriae, and A. meyeri. Bacterial cells were cultured in BHIY broth for 24 h, and 1-ml samples were then collected in micro centrifuge tubes and resuspended at a density of 1.0 McFarland standard (approximately 107 colonyforming units (CFU)/ml) in 1 ml of sterile distilled water. A total of 3.6 μl of the suspension was then used as a PCR template. The detection limit of PCR was assessed by serially diluting known numbers of bacterial cells in sterile distilled water and then subjecting each suspension to PCR. The multiplex PCR mixture contained 0.2 μM of each primer of each group, 10 μl of 2 × Mighty Amp Buffer Ver.2 (Takara Bio Inc., Shiga, Japan), 0.4 μl of Mighty Amp DNA Polymerase (Takara), and 5 μl of the template in a final volume of 20 μl. PCR reactions were performed in a DNA thermal cycler (Applied Biosystems 2720 Thermal Cycler; Applied Biosystems, CA, USA). PCR conditions included an initial denaturation step at 98 °C for 2 min, followed by 30 cycles consisting of 98 °C for 10 s and 70 °C for 1 min. PCR products were analyzed by 2.0% agarose gel electrophoresis before being visualized by electrophoresis in 1 × Tris-borate-EDTA on a 2% agarose gel stained with ethidium bromide. A 100-bp DNA ladder (Takara Biomed, Shiga, Japan) was used as a molecular size marker.
Twenty specific primers covering the upstream regions of the 16S rDNA sequences of oral Actinomyces species belonging to groups A and B were designed in the present study (Figures 1,2). The specific forward primers of group A were designated as MAJF for A. johnsonii, MAORF for A. oris, MANF for A. naeslundii, MAODF for A. odontolyticus, and MAIF for A.israelii, whereas the specific reverse primers were designated as MAJR for A. johnsonii, MAORR for A. oris, MANR for A. naeslundii, MAODR for A. odontolyticus, and MAIR for A. israelii. The amplicon sizes of A. johnsonii, A. oris, A. naeslundii, A. odontolyticus, and A. israelii were 136 bp, 294 bp, 413 bp, 622 bp, and 907 bp, respectively. The specific forward primers of group B were designated as AGEOF for A. georgiae, ADF for A. dentalis, AGERF for A. gerencseriae, AGRF for A. graevenitzii, and AMF for A. meyeri, whereas the specific reverse primers were designated as AGEOR for A. georgiae, ADR for A. dentalis, AGERR for A. gerencseriae, AGRR for A. graevenitzii, and AMR for A. meyeri. The amplicon sizes of A. georgiae, A. dentalis, A. gerencseriae, A. graevenitzii, and A. meyeri were 328 bp, 464 bp, 583 bp, 837 bp, and 942 bp, respectively.

Figure 1: Locations and sequences of species-specific primers for the 16S rDNA of group A oral Actinomyces species. The nucleotide sequence of each primer has been underlined.
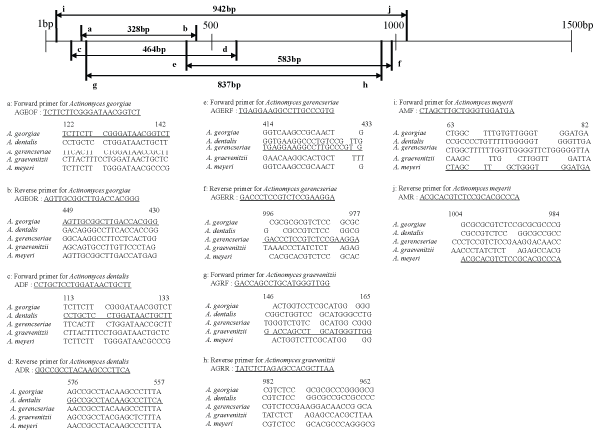
Figure 2: Locations and sequences of species-specific primers for the 16S rDNA of group B oral Actinomyces species. The nucleotide sequence of each primer has been underlined.
Detection limit: Our multiplex PCR method for identifying oral Actinomyces species successfully amplified DNA fragments of the expected size for each species (Figures 3,4). The detection limit was assessed in the presence of titrated bacterial cells, and the sensitivity of the PCR assay was between 5 × 103 and 5 × 105 CFU per PCR template (3.6 μl) for the A. johnsonii-specific primer set with strain JCM 16129, the A. oris-specific primer set with strain ATCC 27044, the A. naeslundii-specific primer set with strain ATCC 12104, the A. odontolyticus-specific primer set with strain ATCC 17929, the A. israelii-specific primer set with strain ATCC 12102, the A. georgiae-specific primer set with strain DSM 6843, the A. dentalis-specific primer set with strain DSM 19115, the A. gerencseriae-specific primer set with strain JCM 12963, the A. graevenitzii-specific primer set with strain DSM 15540, and the A. meyeri-specific primer set with strain ATCC 35568 (data not shown).

Figure 3: Multiplex PCR assay to detect group A oral Actinomyces species.
The primer mixture contained MAJF, MAJR, MAORF, MAORR, MANF, MANR, MAODF, MAODR, MAIF, and MAIR.
Lanes: 1, A. johnsonii JCM 16129; 2, A. oris ATCC 27044; 3, A. naeslundii ATCC 12104; 4, A. odontolyticus ATCC 17929; 5, A. israelii ATCC 12102; 6, A. georgiae DSM 6843; 7, A. dentalis DSM 19115; 8, A. gerencseriae JCM 12963; 9, A. graevenitzii DSM 15540; 10, A. meyeri ATCC 35568; 11, A. viscosus ATCC 15987; 12, Streptococcus oralis ATCC 10557; 13, S. salivarius JCM 5707; 14, S. anginosus ATCC 11391; 15, S. mutans NCTC 10449; 16, S. sobrinus ATCC 33478; 17, Rothia dentocariosa JCM 3067; 18, R. mucilaginosa JCM 10910; 19, Corynebacterium matruchotii ATCC 14266; 20, C. durum ATCC 33449; 21, Neisseria sicca ATCC 29256; 22, Aggregatibacter actinomycetemcomitans ATCC 33384; 23, Staphylococcus aureus JCM 28744; 24, S. epidermidis ATCC 2414. M, molecular size marker (100-bp DNA ladder).
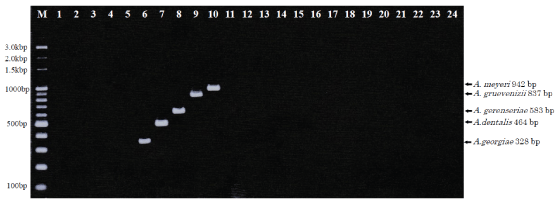
Figure 4: Multiplex PCR assay to detect group B oral Actinomyces species.
The primer mixture contained AGEOF, AGEOR, ADF, ADR, AGERF, AGERR, AGRF, AGRR, AMF, and AMR.
Lanes: 1, A. johnsonii JCM 16129; 2, A. oris ATCC 27044; 3, A. naeslundii ATCC 12104; 4, A. odontolyticus ATCC 17929; 5, A. israelii ATCC 12102; 6, A. georgiae DSM 6843; 7, A. dentalis DSM 19115; 8, A. gerencseriae JCM 12963; 9, A. graevenitzii DSM 15540; 10, A. meyeri ATCC 35568; 11, A. viscosus ATCC 15987; 12, Streptococcus oralis ATCC 10557; 13, S. salivarius JCM 5707; 14, S. anginosus ATCC 11391; 15, S. mutans NCTC 10449; 16, S. sobrinus ATCC 33478; 17, Rothia dentocariosa JCM 3067; 18, R. mucilaginosa JCM 10910; 19, Corynebacterium matruchotii ATCC 14266; 20, C. durum ATCC 33449; 21, Neisseria sicca ATCC 29256; 22, Aggregatibacter actinomycetemcomitans ATCC 33384; 23, Staphylococcus aureus JCM 28744; 24, S. epidermidis ATCC 2414. M, molecular size marker (100-bp DNA ladder).
Assay of representative oral bacteria: As representative oral bacteria, some Streptococcus, Corynebacterium, Rothia, Neisseria, Aggregatibacter, and Staphylococcus were subjected to PCR using the designed primer sets. However, no amplicons were produced from any of the representative oral bacteria (Figures 3,4).
Actinomyces is one of the predominant genera in the oral cavity. At the age of 2 months, one-third of infants are already colonized with Actinomyces [9]. Actinomyces species play a central role in the initial stages of biofilm formation on teeth (i.e., dental plaque) both above (supragingival) and below (subgingival) the gum line [10]. Ten species (i.e., A. naeslundii, A. odontolyticus, A. oris, A. georgiae, A. gerencseriae, A. graevenitzii, A. dentalis, A. johnsonii, A. israelii, and A. meyeri) among the genus Actinomyces are regarded as normal human oral bacteria [3].
Actinomyces species are mainly associated with cervicofacial actinomycosis, oral or cerebral abscesses, caries, root canal infections and periodontitis [11-13]. These microorganisms appear to play a more important role than expected in the pathogenesis of osteoradionecrosis- and bisphosphonaterelated osteonecrosis of the jaw [14-16], and may cause lethal infections such as mediastinitis [17]. As a consequence, fast and reliable identification methods for these microorganisms have become increasingly important.
The isolation and identification of Actinomyces using conventional methods are often difficult and time-consuming. Previous studies were conducted in order to characterize them using phenotypic [18-20] and molecular [21,22] approaches. Most of the available commercial identification kits do not encompass the majority of new species in their database.
The PCR method is a molecular technique that allows species verification at the genetic level, and possesses high sensitivity and bacterial species specificity. It is also useful for the detection and identification of fungi [23]. Moreover, it may efficiently and accurately detect specific microorganisms in clinical samples, regardless of whether they are alive or dead. Also, it might be an innovative method for detection of microorganisms difficult to culture using traditional microbiological techniques. The 16S rDNA region of the bacterial genome provides an ideal target for species identification using PCR [16]. Moreover, multiplex-PCR is a rapid tool that allows for the simultaneous amplification of more than one sequence of target DNA in a single reaction, thereby saving time and reagents [24]. On the other hand, Hirotaki et al. [25] and we [26,27] developed the multiplex-PCR, which allows for the amplification of one sequence of target DNA in a single reaction containing the plural species-specific primers, for bacterial species identification. Previous studies have used different molecular methodologies to identify and differentiate Actinomyces from oral samples after anaerobic cultivation, including PCR-RFLP, chromosomal DNA fingerprinting, 16S rRNA gene sequencing and oligonucleotide-DNA hybridization using universal primers or oligonucletide probes [13,28-30]. Tian et al. reported rapid and sensitive PCR-dipstick DNA chromatography assay that could be read by eye for multiplex and semi quantitative analysis of dental plaque bacteria including Actinomyces species [31].
In silico method helps in designing primers, and there are various programs available for PCR primer design [32]. In the present study, we designed species-specific primers with the already mentioned means, for the identification of oral Actinomyces species using a PCR method. These primers were able to distinguish each oral Actinomyces species and did not display cross-reactivity with representative oral bacteria or other Actinomyces species. Moreover, we developed a multiplex PCR method with the ability to identify and differentiate oral Actinomyces species (i.e., A. naeslundii, A. johnsonii, A. oris, A. odontolyticus, A. israelii, A. georgiae, A. dentalis, A. graevenitzii, A. gerencseriae, and A. meyeri) using only two PCR tubes per sample.
Our multiplex PCR method is easy because the use of Mighty Amp DNA Polymerase Ver.2 (Takara) means that DNA extraction may be avoided, and species identification using this method only takes approximately 2 hours. Thus, the method described herein will allow the prevalence of oral Actinomyces species and their involvement in oral infections to be fully clarified in future studies.
Download Provisional PDF Here
Article Type: RESEARCH ARTICLE
Citation: Kobayashi T, Uchibori S, Tsuzukibashi O, Uezato C, Goto H, et al. (2018) Primer Design for the Identification of Ten Oral Actinomyces Species Using Multiplex PCR. Int J Dent Oral Health 4(1): dx.doi.org/10.16966/2378-7090.249
Copyright: © 2018 Kobayashi T, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Publication history:
All Sci Forschen Journals are Open Access